Lec. 9. Biol C2005/F2401
2006 L.Chasin 10/5/06
© Copyright 2006
Lawrence Chasin and Deborah Mowshowitz Department of Biological
Sciences Columbia University New York, NY
[ATP-synthetase 3-D animation (.mov)]
ATP synthetase
oxidative phosphorylation
substrate level phosphorylation
alternative energy sources:
other sugars
polysaccharides
fat
protein
transamination, deamination
biosynthetic pathways
to monomers
polysaccharide and lipid biosynthesis
protein synthesis ......
Mechanism of ATP synthesis in mitochondria
(ATP synthetase)
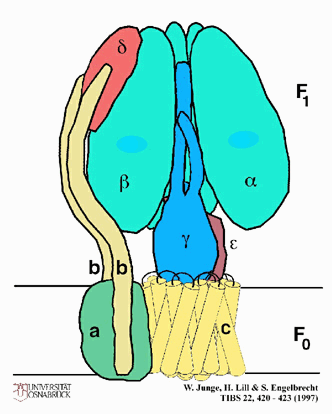 The flow-back is through lollipop-like
structures that populate the inner surface of the inner membrane. See
handout on ATP-synthetase for the structure of this multi-subunit protein
complex. Each lollipop is a complex of proteins; the stem is called
Fo and forms a channel through the membrane.
The sphere is called F1
and contains the ATP SYNTHETASE activity;
that is, it is in the spheres that the generation of ATP from ADP + Pi takes
place. It is the flow-back of H+'s through the F1 spheres that
generates the phosphorylation of ADP by Pi to form ATP.
An analogy would be to use one source of energy to pump water up to a high level
behind a dam (the pumping of protons tied to the free energy released in the
oxidations of the electron transport chain components) and then letting the
water drive turbines to generate electricity as it falls from the high level behind
the dam (the generation of ATP)
The flow-back is through lollipop-like
structures that populate the inner surface of the inner membrane. See
handout on ATP-synthetase for the structure of this multi-subunit protein
complex. Each lollipop is a complex of proteins; the stem is called
Fo and forms a channel through the membrane.
The sphere is called F1
and contains the ATP SYNTHETASE activity;
that is, it is in the spheres that the generation of ATP from ADP + Pi takes
place. It is the flow-back of H+'s through the F1 spheres that
generates the phosphorylation of ADP by Pi to form ATP.
An analogy would be to use one source of energy to pump water up to a high level
behind a dam (the pumping of protons tied to the free energy released in the
oxidations of the electron transport chain components) and then letting the
water drive turbines to generate electricity as it falls from the high level behind
the dam (the generation of ATP)
. This is one place where reading of both texts
can help in the understanding of this very indirect mechanism. Indeed, this
theory was doubted for many years after its proposal by Mitchell (it is also
known as the Mitchell Hypothesis; Mitchell
was known to do experiments in the basement of his mansion, like in the
movies).
The mechanism of this reaction, the ATP
synthetase, has only become clearer in the last few years. The Fo channel
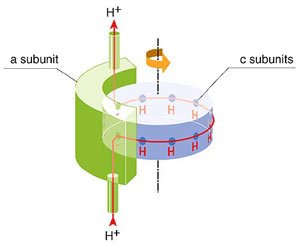
includes 10 c-subunits surrounding a central stalk, the gamma subunit.
A proton outside the mitochondrial membrane flows
back by entering the Fo stem channel where it bind to an amino acids
on one of 10 c subunits comprising the cylindrical channel. This binding
produces a shift in the cylinder by 1/10 turn (see right). As the
c-cylinder turn so does its attached gamma stalk. The top the gamma subunit
reaches into the center of the F1 head structure, and its shape there is in the
form of an asymmetric cam. The F1 head is organized like a pie
divided into 3 identical wedges [Purves
picture]. The wedges can be configured in space to provide a binding site for ADP
and Pi or for ATP. As the cam rotates amidst the alpha-beta subunits of
the F1 head structure, it distorts their structure. Three distinct
distortions are produced and each has an effect on the ability of the alpha-beta
subunits to bind ATP and ADP and Pi on their face that is exposed to the inside
of the mitochondrion. See the
ATP synthetase handout to follow this process.
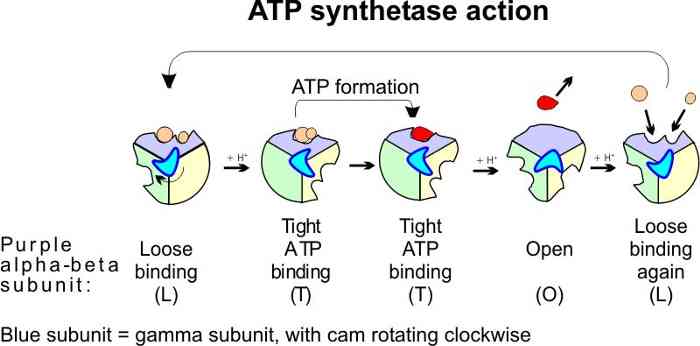
One distortion forces the
ADP and Pi together in one wedge, while the ATP that just had been
formed on another wedge is distorted in the opposite way to release
the ATP. The sequence of these 3 events is thus 1) the binding of ADP and Pi (L), 2)
a kind of mechanical force pushing them together (T), followed by 3) a quick release
of the ATP (O). The formation of these 3 conformations is driven by protons
binding to specific amino acids in the Fo channel. Thus as the protons
flow back into the mitochondrion, the Fo shaft with its cam is spinning.
See a pretty animation of ATP
synthesis from the Website of W. Junge.
To review the action of ATP synthetase
see problem 5-13.
(oxidative
phosphorylation and substrate level phosphorylation)
From glucose, at least .….. But we do not live
from cake alone ….
How about a carbon and
energy source OTHER THAN GLUCOSE?
------------------------------------------------------
Not in live lecture - not responsible for
italicized text:
Where do we get this glucose? Ultimately
from plants, who are able to synthesize it using an energy source obviously
other than glucose, solar energy. I want to now rather briefly turn to
photosynthesis, to summarize what is involved. Just sit back and listen, because
photosynthesis will not be on the exam.
Photo - synthesis, the word and the problem
consists of two parts.
1. Synthesis of glucose from simple CO2 in the
air.
2. Photo: the source of the energy, in the form
of ATP, from sunlight, to carry out this synthesis.
First, the synthesis. Overall:
6 CO2 + 6 H2O --> C6H12O6
+ 6 O2
CO2 is highly oxidized carbon. Need
to add H's from a reducing agent. The ultimate source of these hydrogen atoms is
going to be water, but water is a poor reducing agent (O2 holds its
electrons tightly). So for photosynthesis, we need a source of reducing power as
well as a way to make ATP from ADP.
Overall with ATP:
6 CO2 + 12 NADPH2 + 18
ATP --> 18 ADP + 12 NADP + 1 glucose.
So we need NADPH2 and ATP. Get them both from
the light reactions.
Another piece of cellular machinery: the
chloroplasts, like mitochondria, a membrane delimited cytoplasmic organelle. But they
also contain light-absorbing pigments, including the chlorophylls. When light
energy (one photon) is absorbed by these pigments (each at a characteristic
wavelength), the energy is used to excite a water electron to a higher energy
state.
The excited e- goes down a (different) E.T.C.
ending up at NADP, where a pair of e-'s are accepted to produce NADPH2.
This is the NADPH2 we needed to make glucose.
12 H2O + 12 NADP + 18 ADP + 18 Pi +
60 photons -->
6 O2 + 12 NADPH2 + 18 ATP
+ 18 H2O (from ADP+Pi)
The ATP is made via proton pumping to establish
a hydrogen ion gradient as the electron travels down an electron transport
chain, as in mitochondria.
Efficiency 27%, once e-'s have been excited.
But much less from light overall, as many e- fall back from excited state.
In plants these reaction occur only in chlorophyll-containing tissues (leaves, mainly); other plant tissues use glucose via respiration. And so
do the leaves at night. Glucose is stored, transported, and used for energy.
So plants: CO2 --> glucose + O2
and we (and plants): glucose + O2 --> CO2
The carbon gets shuttles back and forth. In the
planet as a whole, the CO2 is also in equilibrium with vast stores of
inorganic CO2 in the oceans and in the form of carbonates (CaCO2),
limestone.
----------------------------------------------------------------------------------------------------------------
We discussed glucose. But one does not live by
refined sugar alone. Most parts of a big Mac dinner will provide us with energy.
These pathways of degradation of compounds is called CATABOLISM.
The roll:
1. POLYSACCHARIDES:
STARCH = Gluc-gluc-gluc-gluc-....Hydrolyzed to
glucose-1-P, then PhosphoGlucoMutase catalyzes its isomerization to G6P = a glycolytic intermediate . Glycolysis
takes it from there.
See problem 4-13 (4-11 in 16th ed.) Ask yourself, "What steps and enzymes are needed to carry out the reaction shown in the problem: Glucose-P + ATP --> 2 PGAlde?"
Milk shake:
More complex sugars: LACTOSE = glucose-galactose -->
glucose + galactose; galactose then --> --> glucose.
Lettuce:
CELLULOSE --> nowhere (excreted). We can't
handle it (no catabolic enzyme, no cellulase).
The French fry:
2. LIPIDS: fats = triglycerides (structures on
board, refer to handout) = more interesting:
Triglyceride hydrolysis --> glycerol + 3 free
Fatty Acids
For the main Fatty Acid (FA) part of the
triglyceride (see handout):
FA + CoA (ATP [--> AMP] ) to get started -->
FA-CoA --> via FAD (2,3-ene C=C double bond), then HOH addition (OH added distally),
--> via NAD --> C=O
(3-keto, or beta-keto acid, so called
"beta-oxidation", in mitochondria)
+ CoA again (no ATP needed here) --> FA-CoA (smaller by 2 carbons
now), + CH3-CO-CoA = acetyl-CoA = the KC entry compound --> continues
around the Krebs Cycle.
etc. etc. in blocks of 2 carbons. See problem 5-14.
About twice the energy as from glucose, pound
for pound.
How about the glycerol part?
See glycolysis handout. On PowerPoint graphic,
glycerol + ATP --> glycerol-1-P
glycerol-1-P + NAD --> DHAP (= dihydroxyacetone
phosphate) + NADH2
DHAP --> continues in glycolysis (rxn 5). See recitation problems #3, problem 2.
Under aerobic conditions, no problem, the NADH2's
produced will get re-oxidized in the E.T. chain.
Under ANAEROBIC conditions cannot be fermented
(see last lecture).
The burger,
3. = PROTEIN:
Common step for all proteins: hydrolysis of the
peptide bonds by several enzymes (pepsin, trypsin), stomach acid (pH1) denatures
the protein, making it accessible to proteolytic attack --> Amino
Acids.
Subsequent metabolism depends on which of the 20 aa's we're talking
about.
A central reaction is:
Glutamate (R= -CH2-CH2-COOH ) + NAD + H2O
--> alpha-ketoglutarate (--> Krebs Cycle continuation) + NH3
This enzyme is glutamic dehydrogenase (GDH).
Also:
Aspartate: (R= -CH2-COOH) -->
-NH3, --> HOOC-CH=CH-COOH (that's fumarate = a Krebs
Cycle intermediate)
Aspartate deaminase (E. coli but not humans)
The rest of the amino acids cannot lose their
amino group so directly.
So a general problem for aa's: to get rid of the NH2 group: General
solution: Transamination:
aspartate + alpha-KG --> oxaloacetate (OA) + glutamate
(=transamination)
Pass the NH2 to alpha-KG to form glu,
then use GDH to make alpha-KG + NH3 from glu.
Asp becomes OA, another old friend.
Another simple example: ala + a-KG -->
glutamate + pyruvate ( --> KC)
What happens to this ammonia (NH3), sounds bad.
High conc. of NH3 are toxic for many cells (OK for E. coli,
excretes directly and quickly), so combine with CO2 to form urea for
excretion (low urea conc. here, so OK, no denaturation of proteins).
Other catabolic paths are more complex, but the
principle is the same, they get converted to intermediates of the Krebs Cycle or
of glycolysis..
His --> --> --> --> glutamate in humans, but
not in E. coli. So we can do something E. coli can't, for a change (my graduate
work involved this pathway)
So we get energy from protein. Fast for a day
or two, and you start breaking down your own muscle protein to get energy to
keep going. See Problems 5R-4 & 5R-5 for an example of amino acid metabolism.
4. Nucleic acids (in a burger, though not much)
Enough is enough: again, --> KC +
glycolytic pathway intermediates.
Now we are going to turn from the breakdown of
chemicals to the BIOSYNTHESIS of new ones. The
process of building up new compounds from simpler ones is called
ANABOLISM.
1. POLYSACCHARIDES
Glucose: run glycolysis in reverse, but often
with different enzymes for a particular step.
e.g., a phosphatase, uncoupled to ATP for
F-1,6-diP to F6P.
UTP (similar to ATP) + glucose-1-P --> UDP-gluc +
PPi, + glycogen(n) --> UDP + glycogen(n+1)
UDP + ATP --> ADP + UTP
So 2 ATPs are used to put each glucose residue
in glycogen (one to make the G-1-P plus one to regenerate the UTP)).
Also: glucose --> --> gal -->
+glucose --> lactose
All enzymatic, and 1 or a few enzymes for each
polysaccharide, then a repeating structure, so the same few enzymes are used
again and again.
Hyaluronic acid = 2 modified sugars, repeating:
N-acetyl-glucosamine + glucuronic acid:
NAcGA-Glucu-NAcGA-Glucu-NAcGA-Glucu-NAcGA-Glucu - - - -
(see
handout) : glucose --> N-acetyl-glucosamine; glucose --> glucuronic
acid
N-acetylglucosamine + UTP --> UDP-N-acetyl-glucosamine; + growing chain -->
growing chain+1 + UDP
glucuronic acid + UTP --> UDP-glucuronic acid; + growing chain --> growing chain+1
+ UDP
Two alternating enzymes build the polymer from UDP-sugar monomers
2. LIPIDS, phospholipids:
biosynthesis is like degradation, but
reverse, and different enzymes (and not in mitochondria). See
handout.
glucose --> DHAP, (+NADH) --> glycerol-1-P,
+ 2 FAs --> phospholipid; + 3 FA =
triglyceride
Biosynthesis of FATTY ACIDS (see handout)
1) Acetyl-CoA + NADPH2; 2) -HOH step
; 3) NADPH2 again --> repeat in blocks of 2, --> Fatty Acid
NADP is like NAD but with an additional phosphate group. NADPH is usually
used as a reducing agent for biosynthetic reactions, while NAD is used as an
oxidizing agent in catabolic reactions.
3. NUCLEIC ACIDS
Defer, but how about ATP, a NA monomer
glucose --> --> glyceraldehyde-3-P --> ser -->
gly
G6P--> --> ribose-P
ser + gly + ribose-P + NH3 --> +
about 10 steps -->ATP.
(Then ATP <--> ADP many times, for plenty of
energy)
4. Proteins:
Monomers = aa's:
Serine from glycolysis: see
handout
Krebs Cycle --> alpha-keto-glutarate, +NH3
+ NADH2 --> glutamate,
Krebs Cycle --> OA --> transam (via glu) -->
aspartate
Pyruvate, via transamination --> alanine
Isoleucine (ile):
Glucose --> Oxaloacetate --> asp --> asp-P -->
--> --> thr --> --> --> --> --> ile
Added in this path: pyruvate, 2 NADPH2's,
1 NADH2, 2 ATP, transaminations
Made other aa's along the way: asp, thr
Typical biosynthetic pathway: branches,
requires reducing power (NADH2, or NADPH2),
ATP, subject to Feedback Inhibition.
Summary:
SEE OVERHEAD:
YOU ARE WHAT YOU EAT (simplified). Yes and No. Same atoms, but different
compounds. All paths funnel into (catabolism) or fan out from (anabolism) the
central energy metabolism pathways of glycolysis and the Krebs cycle. Due to
coupled reactions or an overall favorable free energy change, these pathways
have Delta Go's that are large and negative.
So that finishes a long section that started
with one important function of proteins, being enzymes. But we
included a discussion of the directionality of the enzymatic reactions, the need
for energy, the metabolic pathways that produce the needed energy, and then
metabolic pathways leading to the needed small molecules in general.
Now let's return to proteins as
macromolecules, and finish off our consideration by seeing how they are
synthesized, how the monomer amino acids are polymerized into polypeptides.
We saw how polysaccharides are made by the
action of a few biosynthetic enzymes, which had as substrates the growing end of
a polymer chain and the free monomer that is being attached to it. So a handful
of enzymes can take care of polysaccharide synthesis. Similarly, a handful
of enzymes can handle the biosynthesis of fatty acids and fats and
phospholipids.
So now let's consider the biosynthesis of proteins in the same
way. Let's start with the synthesis of hexokinase, the first enzyme of
glucose metabolism. Suppose the first few amino acid were met-val-his-leu-gly.
In this case, the growing end is different for each step, so we would need one
enzyme to add val to met, and then another to add his to met-val, and a third to
add leu to met-val-his, and so on. So if hexokinase has 500 amino acids, we
would need about 500 enzymes to put it together. If there are 3000 enzyme
proteins that have to be made this way, then that's 3000 x 500 = 1,500,000
enzymes to do the job. That's a lot of enzymes. And wait a minute: each of these
1,500,000 enzymes is a unique protein itself, and has to be synthesized! So we
quickly see the impossibility of making proteins by a series of simple enzymatic
condensation reactions. We need a new principle, a way of using some
common synthetic apparatus that somehow knows what amino acid comes after what
amino acid in every protein. A mystery until the 1960's. The discovery
of how
this information (what a protein's primary sequence should be) is realized in a
growing cell is one of the greatest achievements in the history of experimental
biology. The answer of course is written in the nucleic acids.
© Copyright 2006 Lawrence Chasin and
Deborah Mowshowitz Department of Biological Sciences
Columbia University New York, NY
 The flow-back is through lollipop-like
structures that populate the inner surface of the inner membrane. See
handout on ATP-synthetase for the structure of this multi-subunit protein
complex. Each lollipop is a complex of proteins; the stem is called
Fo and forms a channel through the membrane.
The sphere is called F1
and contains the ATP SYNTHETASE activity;
that is, it is in the spheres that the generation of ATP from ADP + Pi takes
place. It is the flow-back of H+'s through the F1 spheres that
generates the phosphorylation of ADP by Pi to form ATP.
An analogy would be to use one source of energy to pump water up to a high level
behind a dam (the pumping of protons tied to the free energy released in the
oxidations of the electron transport chain components) and then letting the
water drive turbines to generate electricity as it falls from the high level behind
the dam (the generation of ATP)
The flow-back is through lollipop-like
structures that populate the inner surface of the inner membrane. See
handout on ATP-synthetase for the structure of this multi-subunit protein
complex. Each lollipop is a complex of proteins; the stem is called
Fo and forms a channel through the membrane.
The sphere is called F1
and contains the ATP SYNTHETASE activity;
that is, it is in the spheres that the generation of ATP from ADP + Pi takes
place. It is the flow-back of H+'s through the F1 spheres that
generates the phosphorylation of ADP by Pi to form ATP.
An analogy would be to use one source of energy to pump water up to a high level
behind a dam (the pumping of protons tied to the free energy released in the
oxidations of the electron transport chain components) and then letting the
water drive turbines to generate electricity as it falls from the high level behind
the dam (the generation of ATP)
