C2006/F2402 '02 Key to Exam #2
Q 1-3; each correct answer = 2 pts unless noted. Value of explanation varies and is indicated below.
1. A. Fluorescence will first be seen in the ER and if you add an inhibitor of RNA pol II you will get less fluorescence. Explanation 4 pts. The protein (enzyme E) is part of the endomembrane system, so it must be made on ribosomes attached to the ER, and it must enter the ER co-translationally as it is made. The protein ends up in the Golgi, but it has to enter the ER first. [2pts.] Production of hybrid protein is regulated by the steroid hormone estrogen, so regulation should occur at transcription. In other words, addition of hormone should trigger transcription of the gene for enzyme E. RNA polymerase II is the enzyme responsible for transcription of the gene -- the enzyme catalyzes production of mRNA. If inhibitor is present, you should make less mRNA, and therefore less protein. The less hybrid enzyme the less fluorescence. [2 pts.]
B. Addition of sugars to proteins. (This occurs in ER and Golgi.) The other process mentioned do not occur in the Golgi. The removal of signal peptides occurs in the ER. Degredation of receptors occurs in lysosomes. Destruction of peroxides and toxins occurs in peroxisomes. Addition of NLS occurs as part of protein synthesis in the cytoplasm.
C. Vesicles are moving backwards, trans to medial. (medial to cis was also accepted if explained properly). Explanation 4 pts. These results make the most sense if the Golgi sacs are maturing and moving "forwards" as the proteins within them are modified. Meanwhile, the enzymes that do the modifications are moving "backwards" because they are retrieved from each sac as it moves forward and restored to the next sac as it "moves up." [2 pts.] Since enzyme E works in the medial sacs, it is removed from medial sacs as they become trans and added to cis sacs as they become medial. So depending on how you look at it, the enzyme is transferred backwards from the new trans to the new medial sacs OR the enzyme is transferred backwards from from the old medial (as they move up to become trans) to the old cis (as they move up to become medial). [2 pts. ]
D. Plasmid must have an HRE; chromosomal genes must be making a soluble (intracellular) receptor. Explanation 6 pts. If hybrid protein is produced in response to hormone, the gene for the hybrid protein must be associated with an HRE (hormone response element = binding site for hormone/receptor complex). The HRE must be "cis to" -- on the same DNA as -- the gene that it controls. [2 pts for role/position of HRE.] The cells must make their own receptor for the hormone, using chromosomal genes, as the cells respond to the hormone even if the plasmid is not present. Since the hormone estrogen is a steroid, the receptor should be an intracellular or soluble one -- not a membrane bound receptor on the cell surface, and the receptor should act as a TF. The same receptor (produced by the chromosomal genes) can activate both chromosomal and plasmid genes. [2 pts for role/function of receptor. ] How does addition of the hormone lead to production of hybrid protein? Binding of the hormone to its receptor activates transcription -- the binding may trigger binding of the receptor to the HRE or activate receptor which is already bound.[2 pts for linking roles of receptor and HRE.]
2. A. 400 base pairs, all molecules the same length; 4 molecules of H3/400 BP. (2 molecules of H3 per 200 BP of DNA = per nucleosome.) Bands are multiples of 200 BP and shorter DNA molecules run faster (are closer to bottom). Band X = 2nd from bottom = 2nd smallest = 400 BP.
B. Transcribed region of an active gene. Explanation = 6 pts. Heterochromatin is too tightly coiled to be digested by nucleases -- the linkers are buried and the DNA is not accessible to the enzyme. [2 pts.] Active regulatory regions are extremely sensitive to nuclease treatment as the histones of the nucleosomes have been effectively replaced by TF's. These regions are digested thoroughly by nucleases, not cut at 200 BP intervals. [2 pts.] Active euchromatic DNA is in nucleosomes ("beads on a string" stage) and can be cut in the linkers by micrococcal nuclease to give a "ladder" pattern = multiples of 200 BP. [2 pts.] Transcribed euchromatin is looser than inactive euchromatin, but even transcribed euchromatin is in nucleosomes -- it is not completely digested by nucleases.
3. A. Restriction sites are upstream of the start of transcription (symbolized by the bent arrow); region shown is proximal regulatory region (upstream of core promotor); #112 = base #112, counting from the 5' end of the transcript (= #112 from 5' end of mRNA if no introns in this section). These pictures are always drawn so that transcription goes in the direction of the bent arrow, and the numbers given on the "DNA line" correspond to the positions in the sense strand (with the start of transcription = 0).
B. Exposed in liver cell chromatin; protected only when the gene is NOT transcribed. (1 pt each). The Bam H1 site can always be digested, so it is never protected. The Apa I site can be digested if the DNA is not transcribed, so it is only protected when there is NO transcription.
C. When the gene for enzyme E is not transcribed, Bam H1
site is probably in a linker (neither accepted also), Apa I site is in a core
nucleosome, & area has precisely positioned nucleosomes.
Explanation 6 pts (2 for each part). Note that this region of the DNA -- the
section including the restriction sites -- is a regulatory region, not a
transcribed region. It controls transcription of gene E, but it is not
transcribed itself. Regulatory regions like this are assumed to contain
nucleosomes when the region is "off" but to be nucleosome free when
the region is "on" -- when the gene they control IS transcribed.
The most likely way the Bam H1 site could be accessible to a
restriction enzyme in both transcriptional states is if the chromatin is
relatively loose (both times) and the section including the Bam site is in a
linker between 2 core nucleosomes (when there are nucleosomes present). The
alternative explanation is that this particular spot is never in a nucleosome. To
get full credit for this part of the explanation (C-1, 2 pts) you had to
consider the possibility that the Bam H1 site is in a linker.
The Apa I site could be protected, in the core part of a
nucleosome, when the region is "off," and exposed, without any
histones, when the region is "on." [2 pts.] This fits the data given
and the general assumption that regulatory regions that are "on" are
stripped of nucleosomes. (Note: in the paper from which these results were
obtained, the authors show that the nucleosome is still there when the gene for
enzyme E is transcribed! They show it is still there but is somehow loosened up,
so the restriction enzyme Apa I can get in and digest the DNA inside. They argue
that some "hypersensitive sites" may still have their nucleosomes in
addition to the added TF's needed to start transcription. However the
nucleosomes are in an altered, very loose state-- because of the TF's? -- which
is why the sites are hypersensitive to nuclease digestion.)
The nucleosomes must be aligned (at the same spot in every
DNA in every cell) or the DNA molecules would not always be cut in exactly the
same place. If nucleosomes were positioned randomly, some linkers would be cut
in some DNA molecules and different linkers in others. So you wouldn't cut all
the DNA molecules, from many cells, in the same place. [2 pts.] Q to think
about: How do they know all the DNA molecules are cut in the same place?
Although
we talked about the epinephrine receptor, there are actually several proteins
that are receptors for epinephrine. Each
liver cell has two kinds of epinephrine receptors, called alpha-1 and beta-2.
You culture liver cells in vitro with epinephrine and/or certain
drugs, and you measure the following variables:
(? indicates you forgot to measure this.)
|
Liver
cells incubated with: |
Results |
|||
|
|
Glycogen
phosphorylase |
Cytoplasmic cAMP |
Cytoplasmic
Ca++ |
DAG |
|
epinephrine
+ beta-2 receptor antagonist |
Not
phosphorylated |
No
change |
No
change |
No
change |
|
alpha-1
receptor agonist |
Phosphorylated |
? |
? |
Increases |
|
beta-2
receptor agonist |
? |
? |
? |
? |
|
epinephrine
+ beta-2 receptor antagonist |
Phosphorylated |
No
change |
Increases |
? |
|
epinephrine
+ alpha-1 receptor antagonist |
Phosphorylated |
Increases |
No
change |
? |
4.
Questions A. through G. are based on the experiment described on the last
page. Read the first four questions
before circling your answers. Circle
all correct answers. For some
questions, more than one answer may be correct!
To answer this, you need to know the two pathways we discussed (cAMP, IP3) and to realize that an agonist stimulates a receptor, an antagonist blocks it. Stimulating with an alpha-1 receptor agonist will mimic epinephrine's stimulation of the alpha-1 receptor. This will give the same response as treatment with epinephrine + beta-2 receptor antagonist, since in the latter case only the alpha-1 receptor is stimulated by epinephrine.
(2)
A. Stimulation
of which receptor will lead to an increase in blood glucose?
(alpha-1) (beta-2) (both) (neither)
Blood glucose will increase when glycogen phosphorylase is phosphorylated, since that active enzyme breaks down glycogen to glucose-1-P. This occurs in the last two cases, when epinephrine is given with a beta-2 blocker, so stimulates just the alpha-1 or epinephrine is given with an alpha-1 blocker, so stimulates just the alpha-1 receptor.
(5) B.
Treatment of liver cells with just a beta-2 receptor agonist
will lead to activation of (protein kinase A)
(protein kinase C) (phospholipase C) (glycogen
phosphorylase) (glycogen synthase)
Response to a beta-2 agonist should be similar to response to epinephrine + alpha-1 antagonist, in which case cAMP increases. This is apparently the signal transduction pathway we discussed in class, where protein kinase A and glycogen phosphorylase are activated. Or you could have reasoned that stimulating just the alpha-1 receptor, either with an alpha-1 agonist, or with epinephrine + beta-2 antagonist leads to activation of the IP3/DAG pathway, so you might have used this to conclude that the beta-receptor mediates the cAMP pathway.
(5) (3) C. Stimulation
of either alpha or beta receptors will lead to activation of phosphorylase
kinase, the enzyme that activates glycogen phosphorylase.
But there are two ways to activate phosphorylase kinase:
By phosphorylating it or by binding it to Ca++.
Stimulation of which receptor should lead to Ca++ binding to
phosphorylase kinase?
In
part B., we concluded that beta-2 receptors are the ones stimulated in the
pathway we discussed in class, wherein enzymes were activated by phosphorylation.
So the alpha-1 receptors must be the ones that lead to activation of
phosphorylase kinase by Ca++ binding.
(8) (10) D.
When the alpha-1 receptor is stimulated, you would expect that glycogen
synthase will be (phosphorylated by protein kinase A)
(phosphorylated by protein kinase C) (1) (phosphorylated by protein phosphatase) (dephosphorylated by
protein phosphatase) (activated) (inactivated)
Since there were lots of parts to this question, and since C and D were related, I made part C. worth only 3 points, and part D.worth 10.
List
the steps in the pathway between alpha-1 receptor activation and glycogen.
Diagram just this pathway, not the beta-2 receptor pathway.
Just write the steps that occur in correct order, don’t draw the
molecules.

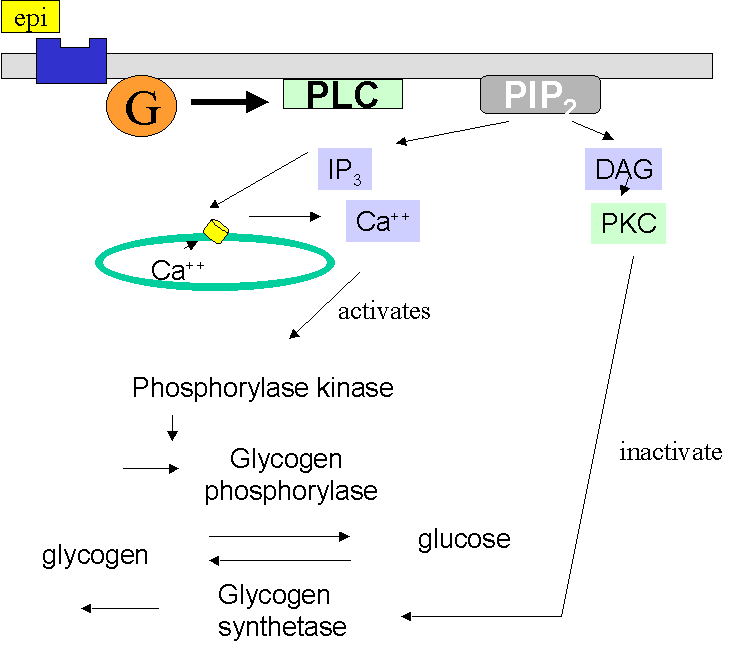
Epinephrine stimulates the alpha-1 receptor, which activates a G protein, stimulating the GTP/GDP exchange (1) which in turn activates Phospholipase C. (1) This catalyzes the breakdown of PIP2 (1) to IP3 (1) which binds receptors on the ER, opening Ca++ channels (1), and DAG (1) which activates protein kinase C. (1) Ca++ binds to phosphorylase kinase, as we saw above, activating it, and it in turn phosphorylates glycogen phosphorylase, which then phosphorylates glycogen to produce glucose-1-P (1) At the same time, we would expect glycogen synthetase to be inactivated, since this enzyme catalyzes the production of glycogen from glucose (1). We know that this enzyme is inactivated by phosphorylation by protein kinase A. Since the pathway here does not involve protein kinase A, but does involve protein kinase C, it is reasonable to conclude that this is the enzyme that phosphorylates glycogen synthetase.
(2)
E. Insulin is a hormone made of 51
amino acids. When insulin stimulates liver cells, the cells increase their rate
of uptake of glucose from the blood. A
short time after epinephrine stimulates alpha-1 receptors in the liver, the
insulin receptors in those cells are phosphorylated.
You would expect that the insulin receptor is being phosphorylated in its
(ligand-binding domain) (DNA-binding
domain) (transcription-activation
domain) (cytoplasmic domain).
Peptide hormones combine with membrane receptors, so no DNA-binding or transcription-activation domains. Hormones bind the ligand-binding domain on the extracellular side, the signal transduction pathways make stuff happen intracellularly, so you'd expect the cytoplasmic domain to be affected.
(5) F. You would
predict that this phosphorylation of the insulin receptor (increases) (decreases)
(2) its ability to bind insulin. Explain
how the information given here lets you reach that conclusion.
Epinephrine
acts to increase EXPORT of glucose from liver to blood, so you wouldn’t expect
insulin to be stimulating IMPORT of glucose at the same time, so the insulin
receptor should be less responsive to insulin.
(2)
G. If both hormones were
administered together to liver cells, you’d expect that (epinephrine has a
permissive effect on insulin) (insulin
has a permissive effect on epinephrine) (epinephrine
and insulin have synergistic effects) (epinephrine
and insulin have antagonistic effects)
More epinephrine causes the cell to be less responsive to insulin, so the two hormones are antagonist, working at cross-purposes.
(5)
A recently discovered hormone is called ghrelin (“ghre” means growth). It stimulates Growth Hormone secretion. Scientists found that ghrelin was secreted by the stomach,
which was a big surprise, since they expected that the major source of ghrelin
would be the (bone) (anterior pituitary) (posterior pituitary) (hypothalamus)
(2)
(liver) (thyroid) (mammary gland) (uterus).
Explain why the tissue you chose was expected to be the source of ghrelin.
One sentence should suffice!
GH
is produced in anterior pituitary
(1), and the release of hormones from there is
generally controlled by releasing or inhibiting hormones from the hypothalamus,(2)
reaching the anterior pituitary by way of the portal vessel, so if ghrelin
stimulates GH release from anterior pituitary, you'd expect it to also come from
the hypothalamus.
Recent
research about ghrelin:
Ghrelin:
an orexigenic and somatotrophic signal from the stomach Nature
Reviews Neuroscience, 2, 551-560 (2001)
Hunger
hormone identified BBC news, Tuesday, 4 December 2001
We discussed how oxytocin travels as a hormone from the anterior pituitary to the uterus, where it stimulates smooth muscle cells to contract. If oxytocin is not arriving at the uterus in the blood (ie, as an endocrine secretion) and it is produced in the uterus itself, then it's likely that it is secreted from some of those smooth muscle cells of the uterus, and it stimulates contraction either in the cell that secreted it or in adjacent cells, ie, it is acting as either a paracrine or autocrine signal.
(2) 6. During early pregnancy, women secrete a hormone
called human chorionic gonadotropin (hCG), which stimulates the gonad to secrete
estrogen and progesterone. Since
all the tropic hormones have a similar structure, hCG sometimes causes
(lactation) (early parturition) (increased
thyroid hormone secretion) (decreased
thyroid hormone secretion)
Tropic hormones include FSH, LH, ACTH, and TSH. If hCG has a structure that is similar to these, and can bind to their receptors, it might mimic their effects. The answer that is most likely to result from stimulation of these receptors is TH secretion , in response to the TSH receptor stimulation.