C2006/F2402 '10 OUTLINE OF LECTURE #9
(c) 2010
Dr. Deborah Mowshowitz, Columbia University, New York, NY.
Last update
02/17/2010 12:28 PM
.
Handouts: Handouts are 9A -- RNA
processing; 9B -- Alternative Processing/Splicing (Not on
web).
I.
Wrap up of Transcription -- Some features not covered
last time. See notes of last time for details
A. Positive vs Negative
Control. See 1st table on handout 8A.
-
Regulation can be "+" or "-" depending on the function of
the protein (& its binding site)
-
Negative control -- If binding of regulatory protein
blocks transcription.
-
Positive control -- If binding of regulatory protein enhances
transcription.
-
Euk vs. Prok. -- Negative control (use of
repressors) seems to be more common in prok.; positive
control (use of activators) more common in euk. (See 1st table on handout
8A)
-
How you tell positive and negative control apart --
by effects of deletions.
B. Basal TF's for RNA pol. II.
1. Terminology: Basal TF's for pol II are called TFIIA, TFIIB, etc.
2. Major one is TFIID; it itself has many subunits. Most studied
subunit is TBP (TATA binding protein -- See Becker
fig. 21-14 (21-15).) Recognizes TATA box when there is one.
3. Other polymerases have TF's too, but TF's for pol II
are of major interest, since pol II →
mRNA
C. Important reminder: Basal TF's bind first to core
promoter, and then RNA pol binds to
them. Takes a lot of proteins to get started. RNA polymerase does
not bind directly
to the DNA.
D. Co-ordinate control. A group of genes can
all be turned on or off at once in response to the same signal (heat shock,
hormone, etc.).
1. Prokaryotes vs. Eukaryotes:
Both
prok. and euk. exhibit co-ordinate control, but mechanism is
different. (See table below.)
2. Location of coordinately controlled genes
(a). In prokaryotes, coordinately controlled
genes are located together in operons.
(b). In eukaryotes, coordinately controlled genes do not need to be near each other -- they just
have to have the same (cis acting) control elements. See Sadava 14.16 (14.14).
3. Control elements:
(a). All genes turned on in the same cell type and/or under the same
conditions share the same control elements -- therefore these genes all respond to the same
regulatory TF's. Result is multiple mRNA's, all made in response to
same signal (s).
(b). Most genes have multiple (cis acting) control elements.
Therefore transcription of most genes is affected by more than
one TF.
(c).
Transcription of any particular gene depends on the combinations of TF's, not just one,
available in that cell type.
4. Differences in TF's.
Different
cell types make different regulatory TF's. Therefore different
groups of coordinately controlled genes are turned on/off. See
Becker fig. 23-24.
5. Comparison of situation in prokaryotes vs
multicellular
eukaryotes:
|
Prokaryotes |
Multicellular Eukaryotes |
|
Coordinately
controlled genes are |
Linked |
Unlinked |
|
Messenger RNA is |
Polycistronic (1 mRNA/operon) |
Moncistronic (1 mRNA/gene) |
|
Operons? |
Yes |
No |
|
Control elements are found |
Once per operon |
Once per gene |
|
Control can be positive or negative but is more often |
Negative -- repressors needed to turn gene off |
Positive -- activators needed to turn gene on. |
II.
Overall Regulation of Eukaryotic Gene Expression
-- What has to be done to make more or less of a protein? A different
protein? What steps can be regulated?
A. If
cells make different proteins,
how is that controlled? If two eukaryotic cells (from a
multicellular organism) make different proteins, what is (usually) different
between them?
Examples: Chicken oviduct cells make ovalbumin -- chicken RBC make
globin*
Human liver cells make transferrin -- human precursors to RBC make globin
*Note: chicken RBC, unlike human RBC, have nuclei
1.
Is DNA different? (No, except in
cells of immune system.)
2. Is mRNA different? (Ans: yes). This means you
can get tissue specific sequences from a cDNA library. (cDNA library =
collection of all cDNA's from a particular cell type.) DNA from each cell type
is the same; mRNA and therefore cDNA is not. See Becker fig. 23-20.
3. Is state of chromatin usually different? (Ans: yes) How is this
tested? Method & result described previously. See figure 23-17
in Becker.
4. Why is the state of the
chromatin different? Is the difference in the cis acting
regulatory sequences or the trans acting factors? (Answer to be discussed in
class.) See Becker fig. 23-24.
5. If mRNA's are different, why is
that? Is the difference due entirely to differences in transcription?
a. Transcription
is different in different cells.
How could it be otherwise?
It could be that all cells transcribe all genes, but only
some RNA's are exported to the cytoplasm and the remaining nuclear RNAs are degraded.
This is not the case. Only selected genes are transcribed in each cell type,
and RNA's from those genes are processed to make mRNA. (For an experiment
that shows this, see figure 23-19 in Becker.)
b. Processing can be different: Splicing and processing of same primary transcripts
can be different (in different cells or at different
times). Different mRNA's (& therefore proteins) can be produced from the same
transcript by alternative
splicing and/or poly A addition. Details & example below.
To review transcription, try
problems 4R-5 and 4R-6A.
B. How can
amount of protein synthesized be controlled? If cell makes more or less protein,
which step(s) are regulated?
1. In prokaryote (for
comparison) -- process relatively simple.
a. Most regulation at transcription.
b. Translation in same compartment as transcription; translation
follows automatically.
c. Most mRNA has short half-life.
2. In eukaryote
-- Gene expression has many more steps & complications than in
prokaryotes -- more additional points of regulation -- not just at
transcription. See Becker fig. 23-11 or Sadava 14.12 (14.11).
a. Transcription is main point of
control, but other steps are often regulated too.
b. Transcription & translation occur in separate compartments.
Translation does not follow automatically.
(1). 2. Transcript must be processed
(capped, spliced, polyadenylated, etc.) -- any of these steps can be
regulated, and there is more than one way to process most primary
transcripts.
(2). mRNA must be transported to cytoplasm.
(3). Translation can be regulated (independently of transcription)
-- can control usage and/or fate of mRNA, not just supply of mRNA. For any
particular mRNA, can regulate 1 or both of following:
(a). Rate of initiation -- can control how often
ribosomes attach and start translation.
(b). Rate of degradation -- can control half
life of mRNA.
c. Different
eukaryotic mRNAs have different half-lives.
Some mRNA's are long lived and some have a very short half
life.
To review regulation, try Problems 4-11 & 4-12.
III. Processing of Eukaryotic mRNA transcripts
Once transcription gets started, what does
it take to get a functioning eukaryotic mRNA?
A. Caps and poly A -- See handout 9A.
Most eukaryotic transcripts that will be
used as mRNA must be modified on both ends (as well as spliced) before they
can be transported to the cytoplasm and used for translation. A "cap" is
usually added to the 5' end and a "poly A tail" to the 3' end. The steps
involved are shown on
handout 9A. (Numbers below match steps on handout.)
(1) Beginning of
transcription.
a. The start of transcription is usually indicated by a bent
arrow.
b. The boxed areas of the DNA = exons; plain DNA between
them = intron.
(2). Capping.
a. A
modified G is added to the 5' end of the transcript shortly after
transcription begins, while the transcript is still being made.
b. The G is added
"backwards" so there is a 5' to 5' connection. (For the curious: The structure of the
cap and how it is connected to the transcript is shown in your texts; see fig.
Becker 21-18.)
c. The cap is represented on the handout as a filled circle.
(3) Transcription continues to
or slightly beyond the end of the gene or transcription unit.
a. There may be no
fixed stop for transcription in eukaryotes (for production of most mRNA); the
addition of poly A (see below) may determine the exact 3' end of the
transcript.
b. Most but not all eukaryote mRNA's contain poly A.
c. Reminder: in
eukaryotes production of rRNA & tRNA are carried out by different RNA polymerases which have somewhat
different properties. these RNA's do not have poly A. (For details see texts.)
(4 & 5). Polyadenylation.
a. A poly A tail -- a string of A's a few hundred long -- is added to the 3' end of
the RNA.
b. Growth of AA...... is 5' to 3' using ATP, enzyme, and splitting off pyrophosphate as
usual. No template is used.
c. The sequence AAUAAA is the signal for the appropriate enzyme to cut
the transcript a bit downstream and add the string of A's. (Downstream = in the
3' direction on the mRNA or sense strand.)
d. Note that the A's on the 3'
end and the G of the cap are not encoded in the template DNA.
e. On the
handout cleavage of transcript = step 4; addition of poly A = step 5. These
two steps may occur simultaneously.
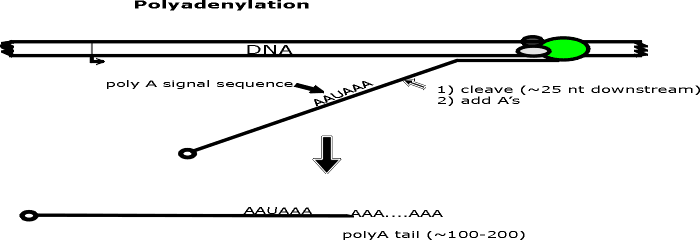
(6) Set up for Splicing. Nothing has happened to the RNA in
step 6 except that it has been labeled to indicate exons and introns (so we
can explain splicing).
a. By the time the transcript is released from the DNA
it already has a cap on the 5' end and a poly A tail on the 3' end. This
RNA -- modified on both ends but not spliced -- is usually called the
primary transcript or pre-mRNA.
b. Some texts refer to unmodified RNA
as the primary transcript, but such a state doesn't really exist, since the
pre-mRNA is modified before it is released from the DNA.
c. The RNA is now ready for splicing (steps 7-9 on
handout). See also Becker fig. 21-22 (21-23) or Sadava 14.10 (14.9).
B. Splicing
of Eukaryotic mRNA
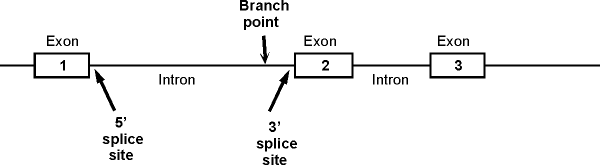
1. A typical picture of a gene with
introns and exons (for reference). The picture below shows a section of
the sense strand of the DNA that includes a gene with 3 exons and 2 introns.
(The picture on the handout has 2 exons and one intron.) Conventions:
-
The picture on the handout
shows double stranded DNA, but genes are often shown as in the picture
below, with only the sense strand actually drawn in.
-
Transcription would start at the 5' (left) end of exon 1 and go to the right.
-
Important features of
intron: Branch point, 5' splice site (also
called the donor site) and 3' splice site (also called acceptor site). These are
shown for the
first intron only. (See also fig. 21-22 (21-23) in Becker or 14.11 (14.10) in Sadava.)
-
Also note that the region to the left of exon 1 is NOT an intron -- it is not
part of the gene. It is part of a spacer in between this gene and the previous
one.
2.
Splicing Details -- See bottom of handout 9A.
a. General Features
(1). Splicing out of each intron occurs in 3 steps (see handout 9A, steps 7-9). At
each step the parts of the transcript are held in place by the spliceosome. The
steps are repeated for splicing of each intron -- many RNA's have many introns. Details are below.
(2). The splice junction at the 5' end of an intron is called the
5' or donor site; the splice junction at the 3' end of an intron is called the
3' or acceptor site.
b. Steps of splicing. See handout 9A at bottom. See also Becker
fig. 21-24 or Sadava 14.11 (14.10). All the steps are catalyzed by the spliceosome. Steps on handout are as follows:
(7) RNA transcript forms loop for
removal of
intron.
(8). Cut at 5' end
of intron
(a). 5' splice site (donor site) is cleaved
(b). loose end of intron (5' end of intron) attaches to branch point in the
middle of the intron, forming lariat-shaped
structure.
(9). Final
Step
(a). The 5' donor site
attaches to the 3' acceptor site, joining the two exons and releasing the intron
in the form of lariat.
(b). The lariat will
be degraded and the nucleotides will be recycled.
(c). The RNA containing the exons
(without the introns) will be transported to the cytoplasm and
translated.
c. N.B: Prokaryotes do not have introns and lack the
machinery needed to remove them.
3. Do exons and translated regions coincide?
See diagram at bottom of
9A. Review from last term:
a.
Exons = sections of genes that are represented in the mRNA.
-
Exons include
untranslated 5' and 3' regions as well as the translated regions.
-
Exons and amino acid coding regions do not coincide, because
there are extra untranslated sections in the mRNA.
-
Exons and mRNA regions do coincide.
b. Exons are not = protein coding sequences, as some texts imply.
(The diagram in Sadava 14.5 (14.4) is incorrect.) Exons include protein
coding sequences, but also include sequences (UTRs) that are represented in the
mRNA but do not code for amino acids.
c. Where are the UTRs?
(1). Leaders. At the 5' end of the mRNA, there is a 5' untranslated region (UTR) or
leader before translation begins (before the first AUG). The DNA coding for
this region is transcribed, and the RNA is not spliced out, but this region of
the mRNA is not translated. The 5' UTR is encoded in one or more exons.
(2). Trailers. At the 3' end
of the mRNA, there is a 3' UTR or trailer that is after the stop codon. The DNA
coding for this region is transcribed, and the RNA is not spliced out, but this
region of the mRNA is not translated. The 3'UTR is encoded in one or more exons.
IV. Regulation at
Splicing -- Results of Alternative Processing
A. There are two ways to get a collection of similar proteins
1. Gene families -- multiple, similar genes exist due to duplication
and divergence of genes. Example: the globin genes constitute a family. Different family
members code for myoglobin, beta-chains, alpha-chains, delta-chains, etc.
Other gene families include the GLUT, SGLT, and IF families.
2. Alternative splicing or processing (See C
below)
-- only one gene, but primary transcript
spliced in more than one way.
B. An example of alternative
processing --
Production of antibody (immunogloblin)
in B cells. See handout 9B and Becker fig.
23-31 -- how to get either soluble or membrane-bound antibody from
alternative processing of the same transcript. (See Sadava 14.21 (14.20) for another
example.)
1. Antibody can be membrane bound
or secreted. Fate of antibody depends on whether peptide has a
hydrophobic sequence near one end or not. Hydrophobic sequence can anchor
the protein in the membrane -- becomes a transmembrane (TM) sequence.
a. If Ab has a potential TM sequence:
Hydrophobic section locks into membrane of ER as protein is made. Vesicle
buds off ER and protein travels through cell as part of vesicle. Protein
remains in membrane of vesicle. When vesicle fuses with plasma membrane, Ab stays in membrane.
b. If Ab has no TM: Ab enters lumen of ER
as protein is made. Vesicle buds off ER, and protein ends up in lumen of
vesicle. When vesicle fuses with plasma membrane, Ab is secreted.
2. Gene has two alternative polyA addition sites. Which
one is used determines final location of protein.
a. Option 1: If one (at end of exon 4/start
of intron 4) is used, protein contains no
hydrophobic potential TM sequence, and protein is secreted.
b. Option 2: If other one (at end of exon 6) is used, protein contains
hydrophobic sequence encoded by exons 5 & 6, and protein stays in plasma
membrane.
3. mRNA can be spliced
and/or poly A added in two alternate ways. Location of protein (antibody) depends on
whether splicing of intron 4 or poly A addition happens
first. Think of it as a competition. Either
a. Poly A adding enzymes get there before the
spliceosome. In that case, poly A is added to site near end of exon 4, and
rest of intron 4 (and rest of gene) is
never transcribed, or
b. The spliceosome gets there first. In that case, Intron 4 is transcribed and spliced out before poly
A can be added. (In this case, poly A is added at the end of exon 6
instead.)
4. Why are 2 forms of antibody needed?
a. Membrane-bound form of antibody:
Serves as receptor
for antigen = trap to detect when antigen is present. Binding of antigen (ligand)
to antibody (receptor) serves as trigger to start secreting antibody.
b. Secreted (soluble) form: Acts as
effector -- carries out major function of immune system -- binds to soluble antigen
in body fluids and triggers destruction of antigen in multiple ways.
C. The
general Principle -- You can get many different mRNAs from
a single gene by the processes listed below. Therefore the number of
possible proteins (the proteome) greatly exceeds the number of possible
genes (the genome).
1. Starting transcription at
different points
2. Ending transcription (adding
poly A) at different points
3. Splicing out different
sections (exons as well as introns) of the primary transcript --
alternative splicing.
To review
regulation &
alternative splicing, try problems 4-13 & 4-14.
V.
Regulation at translation.
A. How to control rate of translation?
In principle:
1. Can regulate half life of mRNA
(control rate of degradation).
a. In prokaryotes most mRNA's have a short 1/2
life; in eukaryotes this is not necessarily so.
b. Different eukaryotic mRNA's have very
different half lives.
c. Note:
proteasomes degrade only proteins NOT RNA's.
2. Can regulate rate of initiation of translation
(control how effectively translation starts).
B. Some
Famous Examples of Regulation
of Translation. (The principles are important; we will
not go into the details.)
1. Use of a regulatory protein:
-- Have a protein that
binds to mRNA (or some other part of the translation apparatus) and affects either initiation and/or degradation, depending on
where it binds. Two examples:
a. Regulation of synthesis of Ferritin &
Transferrin Receptor (& intracellular iron levels).
- Regulatory protein acts like a (prokaryotic) repressor, but
binds to regulatory sequence in mRNA, not DNA.
- Regulatory protein is allosteric, and level of small molecule
effector (Fe) inactivates the regulatory protein.
- The regulatory protein binds to mRNA in the absence of Fe, not
when Fe is high.
- Active form of repressor protein binds to
more than one mRNA. Binds to mRNA for at protein A at 5' end (blocking initiation) and to mRNA
for protein B at 3' end
(blocking degradation).
- This is another example of coordinate control. There is one
trans-acting factor here (repressor), but both mRNA's have the same
cis acting sequence.
- See Becker, figs. 23-33 & 23-34 if you are curious about the details.
A question to think about: Given the information
above, which is protein A, and which is protein B? Which one is ferritin and
which one is the transferrin receptor? Ferritin is an intracellular protein that
stores excess iron. You can check your answer in Becker or using this
diagram.
b. Regulation of globin synthesis by heme
(Becker fig. 23-32). Heme (the prosthetic group of hemoglobin)
stimulates synthesis of globin (the protein part of hemoglobin). In this
case, heme prevents inhibition of translation.
- In the absence of heme, inhibition occurs, and translation is
blocked. No globin produced.
- Heme blocks the inhibition. Therefore, in the presence of heme translation proceeds.
Heme relieves the block in translation, and globin is made.
Interesting features of this case worth noting are:
(1). Inhibition of inhibition results in stimulation; in
other words, (-) X (-) = (+).
(2). Another example of coordinate control. This system ensures
coordination between the supply of heme and of globin. Globin is
useless without heme.
(3). FYI: Heme works by interfering with
phosphorylation -- it blocks a kinase from phosphorylating a
critical translation factor (ElF2 = elongation factor 2).
Phosphorylation of ElF2 (in the absence of heme) inhibits ElF2 and
blocks translation. This is another example of a protein (ElF2) that has
active and inactive forms, and phosphorylation (or dephosphorylation)
interconverts the two forms. (See VI. A below.)
2. Use of a regulatory RNA -- RNA interference (RNAi)
a. Trans acting factors can be RNA. Not all regulatory factors are protein -- some are
short RNA's. (These are usually derived from double stranded RNA -- See Becker figs.
23-35 & 23-36.)
b. How does a short RNA affect translation?
(1). Inhibition (usual case): Small RNA binds to mRNA
→
Formation of double stranded RNA. This triggers degradation &/or
inhibition of translation of the mRNA.
(2). Stimulation: Some recent
cases have been discovered in which small RNA binds to mRNA and 'up
regulates' translation. Mechanism so far unknown.
c. Use in Regulation: Cells naturally produce micro-RNA's that bind to mRNA's and
regulate translation as above. The use of short
regulatory RNA's to block translation appears to be important during
regulation of development. (See Becker 23-36.)
d. Use in the Lab as a tool: Called RNAi = RNA interference. The use
of artificially added short double stranded (ds) RNA to block
transcription/translation and turn genes off is very common. (See Becker
23-35.) Enzymes of cell convert added ds RNA into short single stranded
RNA that interferes with translation and/or transcription as in b. Same
effect as adding antisense RNA (but works better).
VI.
Post Translational Regulation.
Don't forget: regulation occurs after
translation too -- after proteins are made, their activity can be modulated. Many examples
of post translational modification have already
come up and more will be discussed later. Here is a summary (mostly review):
A.
Covalent Modification. Proteins can be modified covalently either reversibly (for ex. by
phosphorylation and dephosphorylation), or permanently (for ex. by removal of
N-terminal met., addition of sugars -- glycosylation, etc.)
See problem 6-3.
B.
Noncovalent Modification. Proteins can be activated or inhibited by reversible noncovalent
binding of other factors -- small molecule allosteric effectors,
other proteins such as calmodulin (an important Ca++ binding protein
to be discussed later), etc.
C.
Degradation. Proteins can be selectively destroyed.
1. Half Lives Vary. Not all proteins
have the same half life.
2. Proteasome: Major factor in
regulation of protein turnover is control of addition of ubiquitin leading
to destruction by proteasome. See Becker 23-38 or Sadava 14.24
(14.22) or the
Nobel
Prizes for 2004.
3. Significance: Important example of
a family of proteins that all have a short half life = cyclins; control
progression through cell cycle. Different cyclins control transitions from G1 to S, G2 to M
etc. Cyclins are made as needed and degraded immediately after use. (Note:
Both mRNA's for cyclins and cyclins themselves are degraded after use. More
on this when we cover the details of the cell cycle.)
D. Location. Proteins
can activated or inhibited by a change of location. For example, transporters
like GLUT4 only work if positioned in the plasma membrane; if they are
sequestered in vesicles they are inactive. Transport of glucose into the cell
can be regulated by moving the GLUT4 in and out of the membrane.
To review post-transcriptional &/or
post-translational regulation, try problem 4-15. By now you should be able to do
all the problems in 4.
Next Time: Intro to
Development: How do you get a multicellular organism with 220 different
cell types?

