 chromosomes, compared to those in a normal karyotype, are diagramed below:
chromosomes, compared to those in a normal karyotype, are diagramed below:1) 24 pts.
Short Answer section. Answer the following as briefly as possible.a) What repeating polypeptide(s) would be produced from the polynucleotide (UCAG)n in a cell free translation system?
Ribosomes could read the message in three frames:
UCA GUC AGU CAG UCA GUC
S V S Q S V
U CAG UCA GUC AGU CAG UC
Q S V S Q S
UC AGU CAG UCA GUC AGU C
S Q S V S Q
Curiously apart from the starting amino acids the peptides from each of the three frames are identical.
b) A mutation that causes
a-thalassemia, a human blood disorder caused by the absence or reduction of the a-subunit of hemoglobin, contains a deletion of two nucleotides just upstream of the AUG initiation codon. The normal version of the gene has the sequence CCCACCATG, whereas the mutant has the sequence CCC**ATG. (* = deleted base)i) Does this particular deletion mutation cause a frameshift, why or why not?
No. A frameshift can only occur in a translated (coding) region. The deleted nucleotides are upstream of the start of translation.
ii) Why do you think this mutation causes
a-thalassemia?The mutation most likely affects the translation of the a-globin message. Mutations just upstream of the AUG codon could easily affect ribosome binding. It could also affect expression or stability of the mRNA. The key point is that the mutation must be some sort of "regulatory" change as the sequence of the protein product is unaffected.
c) Usually there is only a single tRNA species that carries tyrosine. What is its anticodon?
Tyrosine codons are UAC and UAU. The potential tRNA-tyr anticodons are: 3'-5' AUG and AUA.
Since the 3'-5' anticodon AUG can pair with both codons because of wobble pairing and the AUA anti-codon cannot, the AUG anticodon must be the form found in cells with only one species of tRNA-tyr.
1) continued
d) The restriction enzyme Eag I cuts dsDNA at 5' C'GGCCG 3' and Not I cuts at GC'GGCCGC. Would an Eag I cut fragment and a Not I cut fragment have compatible ends? If not why not? If so, could an Eag I fragment ligated to a Not I cut fragment be cut by Not I? If not why not? If so, what percentage of the time?
The ends are compatable. Both cleave to leave a 5' overhand of GGCC.
Eag would leave:
C
GCCGG5'
And Not1 :
5' GGCCGC
CG
These would join to form:
NCGGCCGCN
NGCCGGCGN
The ends could be re-cut by Not I if the leftmost base was a G.
This should happen 1/4 of the time.
h) Explain the differences between a genomic and a cDNA library. Explain how you would use one or the other or both in a chromosome walk.
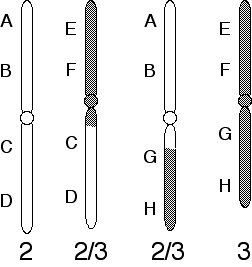
2) 24 pts. A woman seeking genetic counseling, was found to
be heterozygous for a chromosomal rearrangement between the second and
third chromosomes. Her  chromosomes, compared to those in a normal karyotype, are diagramed below:
chromosomes, compared to those in a normal karyotype, are diagramed below:
a) What kind of chromosomal aberration is shown here?
A reciprocal translocation
b) Illustrate how these chromosomes would pair during meiosis. Be sure to label the different segments of the chromosomes.
Consult the text book page 158
2) continued
c) The woman is phenotypically normal. Should this surprise you? Why or why not? Under what circumstance might you expect a phenotypic effect of such a rearrangement?
This is not surprising because the woman is euploid for nearly her entire chromosome complement. The only defective genes are likely associated with the translocation breakpoints and because there are functional copies of each on the intact homologs there should be no phenotype unless the rearranged genes exhibit a dominant effect.
d) The woman has had two miscarriages. She has come to you as her physician and genetic counselor, for advice. It's your task to provide an informed response to her concerns:
i) Is there a genetic explanation of her frequent miscarriages?
Yes, The miscarriages are the result of the production of unbalanced (aneuploid) gametes produced by the normal meiotic process.
ii) Should she abandon her attempts to have a child on her own?
(student answer)
No. It is possible for her to produce an ovum with a normal copy of each chromosome or a translocated copy of each, both of which are viable pairings.
iii) If she continues, is she at risk of having an abnormal child?
(student answer)
No. If she has a child, he/she will be phenotypically normal. But there is a 50% chance that the child, like her mother, will carry two translocated chromosomes, and will thus also have difficulty in having children
3) 10 pts.
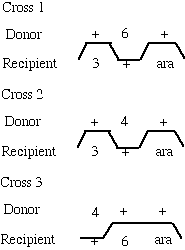
The following data are from 3-factor transductional crosses performed to determine the order of leucine mutations with respect to an outside linked marker arabinose. The donor in each case was ara
+ leu- while the recipient was ara- leu-. Selection was made for leu+ recombinants which were scored for incorporation of the ara+ marker. The data are presented as % leu+ ara+ relative to total leu+ recombinants.|
donor (ara +) |
Recipient (ara -) |
% ara + leu+ |
|
leu-6 |
leu-3 |
12 |
|
leu-4 |
leu-3 |
18 |
|
leu-4 |
leu-6 |
75 |
What is the order of the mutations? (Show your work for partial or full credit.)
The order is ara, leu-6, leu-4, leu-3
Note that there are two types of data--high frequency recombinants and low frequency ones. The high freq class should be the double cross-over progeny and the rare class the quadruple cross-over class. There is only one order that can explain the data:
4) 15 pts.
The first mutants defective in a DNA repair pathway were isolated by the following method: Approximately 107 heavily UV irradiated T1 phage were spread on plates. Then ~5 X 105 mutagenized bacteria were spread on each plate and incubated until colonies arose. Among these colonies were mutants that proved incapable of repairing UV induced lesions in their DNA.(a) Explain the logic of this selection scheme.
(student answer)
The UV irradiated phage had their DNA made non-functional by the radiation. When these phage were used to infect the mutagenized bacteria, any bacteria that were able to repair the UV irradiated phage DNA they were infected with "killed themselves" by allowing the phage to express and lyse the bacteria.
Any bacteria that could not repair the UV-irradiated phage DNA kept the DNA dysfunctional and prevented the phage from growing and thereby survived. Most of the bacteria that survived are the DNA-repari defective mutants we are looking for.
(Note: actually most would be resistant to the phage or otherwise incapable of being infected--nonetheless many DNA repair defective colonies will arise on such plates)
(b) Not all of the colonies that grow up are defective in DNA repair. Explain what other kinds of colonies might grow under this regimen. How would you distinguish the repair defective strains from these other bacteria?
(student answer)
Some of the bacteria that survived could be resistant to infection by the phage (e.g. they do not permit the phage to attach at their surface, or they destroy foreign DNA…)
A good method of identifying the mustants we are looking for is by replica plating, mildly irraidating the bacteria and looking for the ones that died, which were not able to repair their UV-mutated DNA.
5) 15 pts. Although the capture and trading of Great Apes is banned, it is estimated that up to 1000 Chimpanzees are illegally removed from Africa each year. Private owners often disguise this illegal trade by falsifying birth records. To combat this, new techniques are being developed to carry out genetic analysis in Apes. Recently DNA samples were extracted from chimp hair roots and used as templates in PCR reactions. The primers used flank highly polymorphic sites in human DNA resulting from variable numbers of microsatellite repeats. Several suspect chimp offspring and their reputed parents were tested to determine if the offspring were "legitimate" or were the "products of illegal trading." A sample of these data are shown.
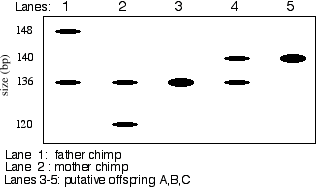
a) Offer an explanation for the origins of the offspring in question. If there are ambiguities state them. Be sure you address whether there are enough data to draw any conclusions, and whether the use of human primers raises any difficulties for the analysis.
Offspring B and C are not the progeny of these parents. Both of them carry a 140 bp allele that neither parent has.
Offspring A could be the progeny of these parents. Chimp A is homozygous for the 136 bp allele. Since both parents are heterozygous for this allele, its quite possible that they are the parents. However, it is still possible that Chimp A is not the progeny of these parents and that the match for this locus is purely coincidental. An examination of other polymorphic loci could resolve any ambiquities.
The use of a human primer should have no bearing on the results. The primers were used on chimp DNA and produced one or two unique chimp bands in each case. This indicates that they bind to only one sequence and that they amplify only the two alleles present at that position. Given the close relationship between chimps and humans it is not surprising that the same DNA sequence flanks the same microsatellite region in both species.
b) What are microsatellite repeats and why can they be highly polymorphic? How many alleles of the repeat locus are present in this group of chimpanzees?
The five chimps carry four alleles for this microsatellite locus. Ones of 120, 136, 140 and 148 bp.
Microsatellite repeats are sequences carrying a variable number of repeats of simple sequence elements such as (CA)n (AAG)n etc. Because they have no function and their repetitive structure facilitates replication mistakes they can exhibit a wide range of sizes within a population. While any one animal carries at most two such alleles the population may carry dozens or more sequence variants at any individual microsatellite locus.
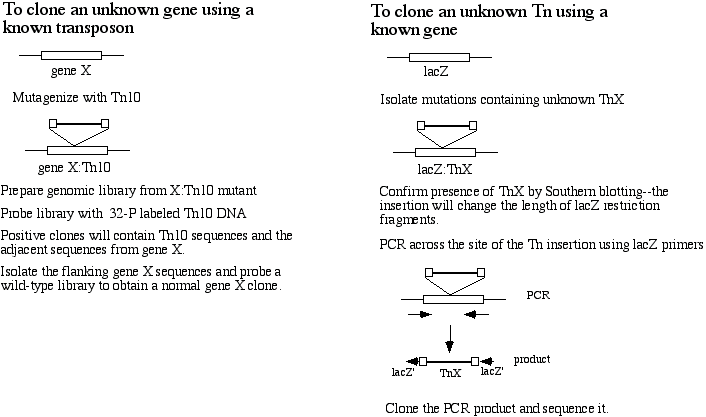
6) 12 pts. When a clone of a transposable element is used as a probe, it is possible to clone gene when nothing is known about the gene's sequence or amino acid product. A similar procedure may be used to clone a transposable element when nothing is known about the DNA sequence of the transposable element. Describe a protocol by which such a transposable element may be cloned.
(Note: If you can't answer this question as written, you may earn partial credit by explaining how you would carry out the experiment implicit in the first sentence.)
Extra Credit 6 pts.
About 20% of all hemophilia A cases in humans are due to an inversion with a break point in intron 22 of the F8C gene. Apparently, this particular inversion has arisen independently many times. Intron 22 is exceptionally large (32kb) and contains a complete small intron-free gene within the intron. This gene, called gene A, has several functional copies upstream from the large F8C gene. The inversion has one break point within the copy of gene A in intron 22 and the other break point is within one of the copies of gene A that lies upstream from the F8C gene. Explain and diagram how his inversion may arise, and explain why this inversion may have arisen many times independently.
Answer will be posted later. Bug me about it in a few days if it doesn't appear.