Study questions for lectures 4, 5 and 6.
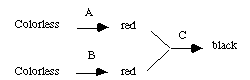
1) A, B and C are independently assorting genes controlling the production of a black pigment. The genes act in a biochemical pathway as follows:

Black can be produced only if both red pigments are present; that is, C converts the two red pigments together into a black pigment. Complete loss-of-function alleles of the genes are designated a, b, c. A homozygous black individual is crossed to a homozygous colorless individual to produce a black F
1. The F1 is selfed. For the F2 generation,
a) what proportion are colorless?
The colorless are genotypes aa bb. (1/4) (1/4) = 1/16 = 4/64.
b) what proportion are red?
p red = Total -(p colorless + p black) = 64 - (27 + 4) = 33/64 are red
Alternatively, red = A_ B_ cc + A_bb + aaBb = (3/4) (3/4) (1/4) + 3/4 (1/4) + 3/4 (1/4)=
9/64 + 12/64 + 12/64 = 33/64
c) what proportion are black?
The black are genotypes: A_B_C_ = (3/4) (3/4) (3/4) = 27/64.
2)
Huntington's chorea is a fatal disease leading to the degeneration of the nervous system. It is found in persons heterozygous for the allele Ht, but has variable expressivity in the sense that a person may develop symptoms as early as childhood or as late as 60 years old, with the mean age of onset at about 40 years.(a) A man with four children (3 female, 1 male) died of Huntington's chorea. The disease is not known in his widow's family. What are the chances that:
i) All four children carry the disease?
(1/2)4 = 1/16
ii) Only the three daughters carry the disease?
(1/2)3 (1/2) = p (3 daughters H) p(1 son w/o) = (1/2)4 = 1/16
iii)Only two children carry the disease?
Apply the sum rule p(HH++) + p (H+H+) + p (H++H) + p(+HH+) + p(+H+H) + p (++HH) = (1/16) (6) = 6/16 = 3/8.
iv) Only one daughter carries the disease?
Apply the sum rule but keep in mind the sexes: Let the first three symbols (H or +) represent females and the last represent the son. p(H+++) + p (+H++) + p(++H+) = 3/16.
(b) In certain families known to carry the Ht allele, the disease is found to "skip" several generations before reappearing. How might this be explained?
Because Ht is dominant and completely penetrant it will "skip" generations only if people do not live long enough to express the phenotype. Since the age of onset can be as late as 60 years this can happen fairly frequently. There is a grey area here between expressivity and penetrance. If you answered incomplete penetrance and explained that this was because people did not live long enough to express the phenotype, that counts for full credit. If you simply said, incomplete penetrance, no credit.
3) For this problem you may need the following background information on Drosophila:
cm = carmine is an eye color mutation that causes dull red "carmine" eyes. cm is located at 18.0 mu on the X chromosome
v = vermilion is an eye color mutation that causes bright red "vermilion" eyes.
v is located at 33 mu on the X. The double mutant cm v has orange eyes.
eag = ether-a-go-go causes flies to shake their legs like crazy when under ether anaesthesia. eag is located at 50 mu on the X.
The telomere of the X is located at 0 mu and the centromere at 68 mu.
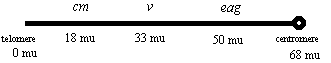
An interesting X-linked recessive lethal mutation was recently isolated in Drosophila. The new mutation, call it lox, for "lethal on the X", originated in a wild-type genetic background. To map lox females heterozygous for lox were crossed to males whose X chromosome is marked with the recessive mutations cm, v, and eag to generate heterozygous females of genotype: lox +++/ + cm v eag. The heterozygous females were crossed to wild-type males with the following results:
|
Phenotypically wild-type females |
2000 |
|
males of genotypes: |
|
|
cm v eag |
700 |
|
+ + + |
3 |
|
cm v + |
110 |
|
+ + eag |
46 |
|
cm + + |
0 |
|
+ v eag |
136 |
|
cm + eag |
4 |
|
+ v + |
10 |
a.) Which gene is lox closest to?
The secret to the problem is to recognize that lox is linked to the wild type alleles of cm v and eag and that any male that receives lox will die. Note that the + + + male progeny are killed (expect for the 3 double XO males). lox is closest to v. lox is most closely linked to the v+ allele. Many fewer v+ progeny survive than any other genotype.
b.) In which interval does lox map? (i.e. Is it between the telomere and cm, between cm and v, between v and eag, or between eag and the centromere?) Explain your reasoning.
lox must map between v and eag.
Progeny that are non-recombinant for the v-eag interval are viable if they are (cm or +) v eag but largely inviable if they are (cm or +) + +. This suggests lox is between v
+ and eag+ or just to the left of v+.Progeny that are nonrecombinant for the cm-v interval are viable if they are cm v (+ or eag) while cm
+ v+ progeny are viable only if also recombinant for v-eag, i.e.+ + eag. This second result is inconsistent with lox mapping to the left of v.If lox maps to right of v the cm ++ class can arise only via a triple XO. This would be expected to be a very rare event and indeed is the only zero class among the progeny. All other rare types could originate with only two XOs.
Consider if lox mapped elsewhere:
If left of cm: 1) the ++ eag class would have required two crossovers and should have been less frequent. 2) The cm ++ class should have been very common. 3) the + v eag class should have been rare since it would require a double crossover. 4) The cm + eag class should have been more frequent that the +v+ class since the +v+ class would require 3 XOs to survive.
If left of v: 1) The ++ eag class would have been the least common type because it would require 3 crossovers. 2) The cm ++ class should have been more common because it would require only a single lox-v crossover. 3) The + v eag class should have been more rare because only cm-lox recombinants would survive.
If right of eag: 1) cm v + class would be rare since it would require a double XO to survive. 2) ++ eag class would have been very common. 3) cm ++ class would be more abundant since double XO freq ~ (.17)(.15). 4) + v+ class should have been rarer than cm + eag since it would require 3 XOs to generate the progeny.
c.) Calculate the map position of lox.
Total v-eag recombination events = [+++ ](3)2=6 + [cm v +]110 + [++eag] 46 + [cm+eag] 4 +
[+v+] 10 = 176
v-lox recombinants = 3 + 46 + 4 = 53
lox-eag recombinants = 3 + 110 + 10 = 123
53/176 = 0.301 of the total v-eag crossovers occurred between v and lox.
lox is located at 30.1% of the distance from v-eag = (.301) (17 mu) = 5.1 mu to the right of v at 38.1 mu.
Alternatively 123/176 = .699 of the v-eag crossovers occurred between lox and eag allowing lox to be placed at (.699) (17 mu) = 11.9 mu to the left of eag at 38.1 mu.
4) Explain how balancer chromosomes could be used to stably propagate an autosomal recessive lethal mutation if the balancer chromosome is homozygous viable but carries a recessive sterile allele that eliminates both ovaries and testes. Assume the balancer is marked with a dominant mutation Ha that produces flies with many extra hairs. What phenotypes, and in what proportions, would you expect in the stable stock?
The stock is maintained because only fertile animals have the genotype lethal/ Balancer. In the population 2/3 will be fertile extra hairy animals, and 1/3 will be sterile extra hairy animals.
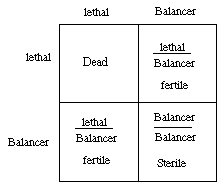
Because the only fertile animals are lethal/ Balancer, the matings and ratios of gametes and progeny are always the same.
(b) If you want to stably propagate an X-linked lethal mutation with a balancer chromosome containing a recessive sterile allele, would you use a female-specific sterile, a male-specific sterile, or a non sex-specific sterile mutation?
You would have to use a female-specific sterile mutation. A male-specific sterile would leave the males either sterile or dead. The same with a non-sex-specific sterile. Only a male fertile and female-sterile balancer will maintain the stock.
All productive matings: lethal/ balancer X balancer/ Y
5
) Both hemophilia A h(A) and red-shift color blindness (r) are X-linked recessive traits. The h(A) and r genes are separated by 4 cM. In the pedigree shown, what is the probability that the individual male (IV-1) is color blind but not affected by hemophilia?
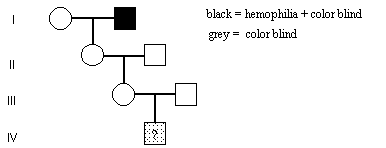
The desired genotype is + r / Y. This could occur if female III-1 were genotype h(A) r / ++ or if she were genotype + r /++. Thus the probability that IV-1 is + r /Y is equal to the sum of p(of son being + r if mother was h(A) r /++) and the p(of son being + r if mother was + r /++ ).
Female II-1, is genotype h(A) r /++
Female III -1 is either genotype h(A) r /++ (p = 1/2(.96)) or
genotype + r /++ (p = 1/2 (0.04))
So probability of son being + r is:
= (1/2(.96)) (1/2(.04) + (1/2 (0.04)) (1/2) = 0.0196