1)
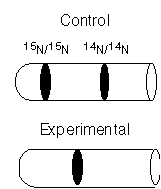
A meteorite landed on earth carrying with it a sample of extraterrestrial bacteria. Homer Simpson was assigned the task of determining the mechanism of DNA replication in this organism.Homer grew the cells in unlabeled medium and then transferred them to 15N containing medium for exactly one generation. He extracted the DNA and subjected a portion of it to CsCl density gradient centrifugation. The banding pattern is shown below with reference to control DNA isolated from cells grown in 14N or 15N media.
(a)
Homer claims to have demonstrated that extraterrestrial DNA replicates in a semiconservative manner. Why is his claim premature?
The dispersive mode of replication is also consistent with this result.
(b) Unfortunately, Homer used all of the bacteria to extract the DNAs and the organism is now extinct on earth--D'oh! Devise an experiment using the leftover DNA samples and CsCl centrifugation, that would distinguish between semiconservative replication and the remaining alternative mode(s) of replication.
To distinguish between dispersive and semiconservative replication one could do the following experiment: Denature the experimental DNA and analyze the single strands (ss) in a CsCl density gradient . If replication is semiconservative, you should two bands, one of ss 14N density and one of ss 15N density (relative to denatured control DNA). If replication is dispersive you should get a band of ss DNA at an intermediate (mixed 14N and 15N) density.
No credit
was given for answers that required further growth of the bacteria--The bacteria were EXTINCT!!!
(At least 1/3 of the answers on last years exam fell into this catagory!)
2)Temperature sensitive mutations are useful for analyzing essential biochemical pathways. A variety of ts mutations affecting DNA synthesis have been isolated in E. coli. Assume that the proteins affected by the following mutations function normally at 30˚ but are inactive at 42˚C.
dnaB
ts (helicase)dnaE
ts (pol III polymerase function)dnaG
ts (primase)dnaQ
ts (pol III 3'-5' exonuclease function)lig
ts (DNA ligase)polA
ts (DNA polymerase I: Polymerase function, 3'-5' exonuclease, 5'-3' exonuclease)
(a) Which of the mutants would be expected to accumulate Okazaki fragments after shift from 30˚ to 42˚ C?
ligase and polA mutants should accumulate Okazaki fragments since replication continues in both situations but the discontinuously synthesized fragments cannot be joined. In the case of ligase because there is no ligase, in the case of polA it's because the RNA primers are still in place and the fragments cannot be ligated.
(b) DNA mutants can be categorized by whether they lead to a rapid (within seconds) or a slow (20 min - 1hr) cessation of DNA synthesis after shift to non-permissive conditions. (DNA synthesis is measured by incorporation of radioactive thymidine.) Which of the mutants would produce a fast-stop phenotype and which a slow-stop phenotype? If you think some of your answers may be ambiguous state why.
dnaB
ts --fast stop. If you can't separate the strands replication will halt immediately.dnaE
ts --fast stop. Pol III replicates the chromosomes. W/o it, no incorporation.dnaG
ts --fast or slow stop accepted. Best answer is probably slow stop since one would expect that continuous strand synthesis would proceed until replication was finished. Discontinuous strand synthesis would halt quickly. In real life primase mutants are fast stop. Probably because the Pol III replication complex (Fig 11-30 in Griffiths) falls apart.dnaQ
ts -slow stop--ambiguous--may not stop at all. This is the error correcting function of pol III replication should continue without much change but the error rate would be expected to go up.lig
ts --slow stop. Synthesis continues but Okazaki fragments cannot be joined.polA
ts -slow stop. Chromosomal replication continues, but primers are not removed and replaced with DNA.
(c) A mutation called mutD (for "mutator gene D") is allelic to one of the mutations listed above. The mutD phenotype is manifested as an ~ 10
5-fold increase in the rate of spontaneous mutation. Which of the genes listed above is most likely to be altered by the mutD allele? Why?
mutD
is allelic to dnaQ. Removing the editing function of pol III should increase the error rate by an enormous amount. polA is not as good a choice since Pol I is responsible for replicating only a tiny fraction of the chromosome in each replication cycle.
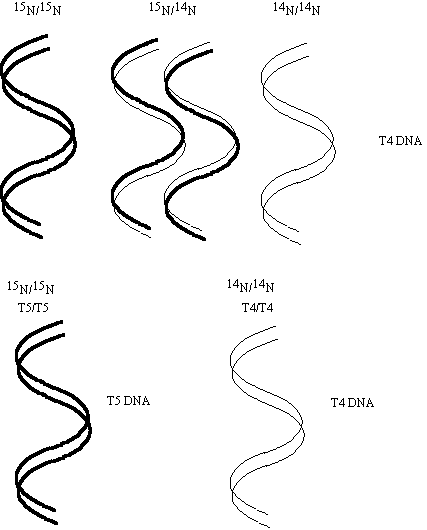
3) When DNA from bacterial virus T4 grown in 14N medium, is denatured and mixed with denatured DNA from T4 that was grown in 15N medium and allowed to renature three bands are observed in a equilibrium density gradient centrifugation experiment. When denatured 14N T4 DNA is mixed with denatured 15N DNA from bacterial virus T5 and allowed to renature only two bands are observed. Explain these results.
The T4 DNA forms three bands in a density gradient because both
14N and 15N strands can associate to form double stranded molecules containing either two heavy, two light or one heavy and one light strand. When light T4 DNA is mixed with heavy T5 DNA only two bands form since T4 DNA will not pair with T5 DNA because the sequences are not complementary.