Note
the exam was worth 102 pts. Who says exams have to be worth 100?
1) 12 pts. Construct pedigrees for traits that could not be:
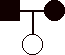
a)
autosomal recessive
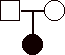
b)
autosomal dominant
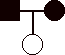
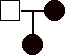
c)
sex-linked recessive
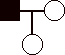
d)
sex-linked dominant.
2) 15 pts. Answer
True or False. If the statement
is TRUE, explain or give a supporting example. If the statement is FALSE,
either explain the problem with the statement or give a counter example.
a) Wobble pairing allows
one form of tRNA for some amino acids to be used at several of the codons for
that amino acid.
True. Wobble pairing reflects the relaxed
base pairing specificity between the third base in a codon and a tRNA. Thus a tRNA with the anticodon 3’
GAU 5’ could pair with either the CUA or CUG leucine codons.
The most common mistake was to assume that the wobble is the same as code degeneracy. Degeneracy refers to the existence of multiple codons for the same amino acid. Wobble refers to the ability of some tRNA species to recognize more than one codon for an amino acid.
b) Cells that are missing
chromosomes are aneuploid.
True. Euploid cells have a normal chromosome
complement. Aneuploidy reflects
deviations from that such as loss or addition of chromosomes.
c) The principle of
segregation refers to the separation of the two alleles of a gene into
different gametes.
True. This is Mendel’s First Law.
d) In birds, butterflies,
and moths the Z chromosome is a sex chromosome and the W chromosome is an
autosome.
False. The Z and W chromosomes are both sex
chromosomes. The W being
approximately equivalent to the mammalian Y.
e) Penetrance can be seen as an extreme
form of expressivity, where some individuals with the mutant genotype do not
show a phenotype at all.
True. Expressivity refers to the degree to
which a phenotype is expressed. If
expressivity was so extreme as to lead to the occasional case where the mutant
phenotype was not expressed it would be equivalent to variable penetrance.
3) 21 pts.
Short Answer section: Answer
the following questions with a short explanation, simple diagram or by the
appropriate calculations.
a) The chromosome of Mycobacterium
tuberculosis contains 18%
adenine.
i)
What is the percent G+C in the genome?
64%. 18% A, 18% T, 32% G, 32% C.
ii)
What is the percent U in the mRNA?
You can’t tell
as the base content of the mRNA may not precisely reflect that of the genome,
as not all sequences are transcribed into mRNA. Most likely, however, it will be fairly close to the % T in
the genome so we accepted 18% U as an answer. If this were a eukaryotic organism, with introns and large
amounts of non-coding sequences, we would not have accepted a % figure.
b) During methyl-directed
mismatch repair, why is it necessary to distinguish between the template strand
and the newly made daughter strand?
How is this accomplished?
So that the
newly-made mistake can be corrected based on the original correct template
strand. This is accomplished by
adding methyl (-CH3) groups to certain nucleotide residues of DNA
after replication. Thus newly
synthesized DNA is unmethylated.
c) In bacteria, incorrect
amino acids are inserted during translation at a rate of approximately 10-3
(one incorrect amino acid per 1000 incorporated). If an incorrect amino acid is inserted
into a crucial position of an essential protein, what will the consequences be
for the cell? Explain.
None,
since the cell will produce many molecules of an individual protein the
overwhelming majority of protein molecules will be unaffected and thus fully
functional. One mutant protein
molecule is effectively diluted out. Remember translation by one ribosome is independent of
another. The mistake is not at the
DNA level and thus is not heritable.
An aside, some eukaryotic proteins, such as dystrophin
(mutated in some forms of Muscular dystrophy), are so large that it is unlikely
that any single protein molecule will have the exact sequence specified by the
DNA given the relatively high translational error rate.
d) Honey bee males are
haploid. Would you expect them to
produce sperm via a mitotic mechanism or a meiotic mechanism? Explain.
Meiosis
is a reductive division. Mitosis preserves the chromosome complement. Since the males are already haploid
there is no need for a reductive division to generate haploid sperm.
e) A balding man
undergoes a treatment which transfers some of the hair from the sides of his
head, where it is still plentiful, to the top. Is he altering his phenotype,
his genotype, or neither? What if the hair comes from another head?
In the first, case
genotype is unaltered, but the phenotype (appearance) is.
In the second, case
the hair comes from another genotype so both the genotype and phenotype have
changed. The genotypic change is somatic and limited to the transplanted cells.
The man's own cells remain unchanged genetically.
Note: The question referred to the man's genotype, which changed because his appearance changed. If we discussed the phenotype of the individual hair cells, the phenotype exhibited by the cells would be unchanged.
I stole this question from another text book. Upon reading your answers I decided that many alternative answers were acceptable. Basically if your thinking was clear and your argument cogent you received full credit.
f) A seed dealer wants to sell three
different packs of snapdragon seeds that will produce either red, white, or
pink flowers. Explain how this
could be done.
Homozygous RR plants will produce seeds that produce red
flowers.
Homozygous rr plants will produce seeds that produce white
flowers.
Crossing RR and rr plants will produce seed that will develop as
pink flowers because the R
allele is incompletely dominant to r.
4. 12
pts. Shown below is a snapshot of
the DNA during replication. Answer
the following questions, referring to the bold-faced letters when nececssary.
![]()
5’…A G C C T A T T A G G G C G A T A A A T G T C T C C … 3’
DNA Pol III T C G G A T A A U C C C G C U A U T T A C A G A G G...
![]()
D
C
B
![]()
![]()
![]()
a) In which direction is DNA Pol III traveling (left or right)?
Left
b) Between which two nucleotides does a covalent bond not exist (A-D)?
D
c) Between which two nucleotides will DNA Pol I initiate (A-D)?
D
d) Between which two nucleotides could a pyrimidine dimer form (A-D)?
A or D.
5) 16 pts. A human hemoglobin variant, "Hemoglobin Wayne", exists and causes a mild anemia when homozygous. Normal a-hemoglobin is 141 amino acids long, while the Wayne
a polypeptide is 146 amino acids long.
a) Describe how this
phenotype could arise either via a single-base substitution or a frameshift
mutation.
A
frameshift mutation late in the coding sequences would change the frame of the
normal termination codon and translation would continue until it reached a stop
codon in the new reading frame.
A single base
substitution that changed the normal stop codon to a sense codon would result
in a longer protein since translation would continue beyond the normal stop
site until a new nonsense codon was reached.
Beginning with the 141st (last) codon (Arg) of the a-globin gene, the 3' end of the wild-type
a globin mRNA has the sequence:
5'..CGU UAA GCU GGA GCC
UCG GUA GCA GUU CCU CCU GAC AGA UGG GCC UCC CAA CGG GCC CUC CUC CCC UCC UUG CAC
CGG CCC UUC CUG GUC UUU GAA UAA AGU CUG AGU GGG....3'.
b) Based on this information
how was the Wayne allele generated?
If the normal stop
codon were changed to a sense codon translation would continue until a new
in-frame stop codon UAA is reached.
This would cause the addition of 31 C terminal amino acids, not the 5
present in the Wayne hemoglobin.
There
is potential stop codon in the -1 frame located 14 nucleotides downstream of
the normal stop. If a single base
were deleted late in the globin coding sequence the reading frame, starting in
the normal stop codon, would be NNU AAG CUG GAG CCU CGG UAG. This would result in a 5 amino acid
addition which is the Wayne phenotype.
c) What is the sequence of the four
C-terminal amino acids of the Wayne a-hemoglobin polypeptide?
L
E P R
CUG GAG CCU CGG
6) 14 pts. Recently the papers reported the cloning of a domestic cat. The donor cat was a calico—a
mixture of orange, and dark tabby-striped fur with large white areas. The clonal offspring “CC”
(“Copy Cat” or “Carbon Copy”) is a dark tabby with
white spotting.
a) If the experiment were repeated several times
with different cells from the same donor, to generate “Xerox”,
“Canon”, “Stallone”, and “Lethal Weapons”
what progeny phenotypes would you expect to observe?
Due to random
X-inactivation, the donor, which was heterozygous for the X-linked O and o
alleles, expresses a mixture of orange tabby markings in the cells which
inactivated the o-bearing X (express O) and dark tabby markings in the cells
that inactivated the O-bearing X (expressing o). The phenotype of CC indicates that X-inactivation is
maintained during cloning. That
is, the X that was active in the donor cell is active in all cells of the
recipient. Thus CC’’s
siblings should be either orange tabby and white (white is due to the Piebald
locus) or tabby and white like CC.
None should look identical due to differences in the variably expressed
Piebald spotted phenotype.
b) If X-inactivation were reversed upon
transplantation of a donor nucleus into an egg and then re-established during
embryonic development, would “CC” have looked identical to her
genetic mother? Why or why not?
If X-inactivation
were reversed CC would have been a Calico as she would have expressed the O
allele in some cells, the o allele in others, and she would have expressed the
Piebald phenotype. She would not
have looked the same as her donor mother because the random process of
X-inactivation would have generated a different orange and dark tabby pattern
and because her pattern of white spotting would have been unique due to
variable expression of the Piebald phenotype.
7) 12 pts. This pedigree describes inheritance of a rare
autosomal disease in two unrelated families. Couple III.2 and III.3 would like to have children. What is the probability that, if they
have two kids (IV), neither of them will be affected by the disease?
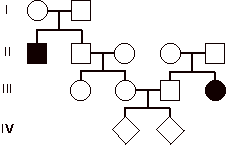
Note: This problem was somewhat more
difficult than I realized when I put it on the exam. If you made certain kinds of incorrect assumptions at the
beginning, it would have been very difficult to identify your error. Accordingly, we have accepted some
wrong answers for full credit.
Please check the answers below carefully to see how to answer the
problem.
The preferred approach
to answering to this problem was:
P (II-2 is heterozygous) = 2/3
P (III-2 is heterozygous) = 2/3 X 1/2 = 1/3
P (III-3 is heterozygous) = 2/3
P(both parents are heterozygous) = 2/9 (thus, P at least one parent
homozygous = 7/9)
P(both kids unaffected) = (P[both kids unaffected if
parents heterozygous] + P [at least one parent is homozygous])] =
(2/9 X 3/4 X 3/4) + 7/9 = 18/144 + 112/144 = 130/144 =
90.28%.
An alternative means of arriving at the correct answer
is:
P (II-2 is heterozygous) = 2/3
P (III-2 is heterozygous) = 2/3 X 1/2 = 1/3
P (III-3 is heterozygous) = 2/3
P(both parents are heterozygous) = 2/9
P(both kids unaffected) = 1-(P[only the first kid is
affected] + P[only the second kid is affected] + P[both kids are affected]) =
1 - ([2/9 X 1/4 X 3/4] + [2/9 X 3/4 X 1/4] + [2/9 X 1/4
X 1/4]) =
1- (6/144 + 6/144 + 2/144) = 1 – (14/144) =
130/144 = 90.28%
We
also accepted the following incorrect answer for full credit. However, you should understand where
you went wrong.
P (II-2 is heterozygous) = 2/3
P (III-2 is heterozygous) = 2/3 X 1/2 = 1/3
P (III-3 is heterozygous) = 2/3
P(both parents are heterozygous) = 2/9
P (IV-1 is affected if parents heterozygous) = 1/4.
Overall, P(IV-1 is affected) = 2/9 X 1/4 = 2/36 = 1/18. (Same P for IV-2)
P (IV-1 is unaffected) = 1-P(IV-1 is affected). (Same P for IV-2)
So P(IV-1 is normal) = 17/18.
Thus P(IV-1 and IV-2 are both normal is 17/18 X 17/18 =
289/324 = 89.2%.
Here’s the mistake.
The P(both kids
unaffected) is actually = 1-(P[only the first kid is affected] + P[only the
second kid is affected] + P[both kids are affected]).
This is
not the same term algebraically as used immediately above.