1) 32 pts.
Answer True or False. If
the statement is TRUE, explain or give a supporting example. If the statement
is FALSE, either explain the problem with the statement or give a counter
example.
a) A testcross can be a simple tool to determine if two genes are linked.
True. A testcross (a+/+b X ab/ab) produces progeny phenotypic ratios of 1:1:1:1 if the genes are unlinked. Deviations from the 1:1:1:1 ratio are very easy to detect and indicate linkage
b) The likelihood of two independent crossover events can be estimated accurately by multiplying the individual probabilities of two crossover events.
True,
but I’d intended it to be false. As written, it is literally true, since the crossovers
are independent, but in the real world crossovers are not really
independent due to interference, which makes it impossible to calculate double
crossover frequency from map distances alone.
(Answers graded accordingly. (That’ll teach me to write exams at 1:00 AM the night before a conference!)
c) Enhancer mutations exaggerate the effects of mutations in another gene.
True. Enhancer mutations increase the expression of the mutant phenotype of another gene. En(wa), for example, lightens the eye color of wa mutants thus making it more mutant and less wild-type. This should be distinguished from mutations in an enhancer element.
d) In complementation, mutations that complement are allelic.
False. Complementing alleles are in different genes and thus are not allelic.
e) 1 centi Morgan (cM) = 1/100 map unit = 1% recombination.
False. It should read 1 map unit.
f) A trp- auxotroph will be unable to grow on glucose minimal medium supplemented with tryptophan.
False. A trp- auxotroph requires tryptophan to grow. Since trp is on the plate it will grow.
True/false continued…
h) Conjugation, but not transduction or transformation requires DNA synthesis for transfer.
This question was not graded.
i) The number of copies of a given gene can be determined with a Northern blot.
False. The number of copies can be determined using a Southern blot. Northern blots contain RNA samples. The intensity of the hybridization signal reflects RNA abundance but not gene copy number.
j) A shortened version of a gene can be detected by Southern blot.
True. A shortened restriction fragment will migrate more rapidly in the gel than will the wild-type fragment.
2) 10 pts. Onion bulbs may be red, white or
yellow. These color differences
are under the control of two pairs of genes that interact epistatically. The F2 generation of a cross produced
40 yellow, 50 white and 110 red onions.
a) give a generalized genotype for all
three onion bulb colors
A_,
B_ = red
A_,
bb =
yellow
aa,
B_ and aa,
bb =
white.
b) Give a possible mating sequence that
would produce the F2 generation above.
AABB X aabb
AaBb X AaBb
=
9:3:3:1 = 9 red, 3 yellow, 4
white. = 110, 40, 50.
3)
6 pts. A man is heterozygous for a disease gene D/d and also for a microsatellite repeat (ms) linked at a distance of 7 cM. The chromosomal arrangement is D
ms(33) / d ms(27). What percentage of his sperm will be D
ms(27)?
1/2 the sperm will carry the D allele. Of those 7% will have recombined between the D locus and the ms locus. Thus, 1/2 X 0.7 = 3.5% of sperm will be D ms(27).
5) 12 pts. The normal sequence of genes and centromere for a particular chromosome is:
![]()
a) What aberration is present in each of the following chromosomes?
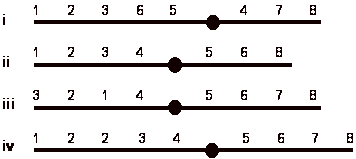
i)
pericentric
inversion
ii)
deletion
iii)
paracentric
inversion
iv)
duplication.
b) If crossing over occurs between the 2 and 3 genes in a iii/normal heterozygote, what genes will be contained on the acentric chromosome fragment that is produced?
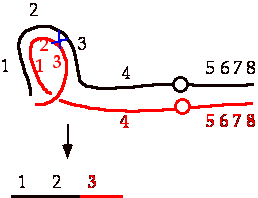 One, two, and three.
One, two, and three.
6) 10 pts.
a) Given the
following double stranded DNA template,:
5’
CTTCGAAATTC….GENE 1….TCTCCCGATCGG….GENE 2….AAGATCAAATCCTTTGCTCT
3’
Which two primers
(written 5’à3’)
would you use in a PCR reaction to amplify only Gene 1?
|
a)
TTCGAAATTC |
b)
CCTTTGCTCT |
c)
TCCCGATCGG |
|
d)
GAATTTCGAA |
e)
GATCGGGAGA |
f)
AATCCTTTGCT |
a) and e).
ON the sequence above, a) is the forward primer and e) the reverse
primer.
b) Primers used for PCR (circle ALL that apply):
i) Are complementary to part of the
template
ii) Are single stranded
iii) Are composed of dNTPs
iv) Will be present at the 3’ end of
the newly synthesized DNA molecule after a round of PCR
i and ii.
7) 14 pts. Suppose a gene has the following restriction map:

Given two plasmids pCU20
and pCU45 (only cloning restriction sites are shown):
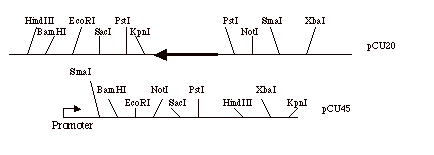
a) Which restriction
enzymes can you use to move the gene from pCU20 to pCU45 so that the gene is
transcribed from the plasmid promoter?
(Diagrams of pCU20 and pCU45 above are not to scale. The polylinker regions are greatly
expanded relative to the inserted gene.)
Xba I
+ Kpn I
or
Not I
+ Hind III
or
Not I
+ Kpn I
or
Pst I
+ Kpn I
b) Which enzyme could you
use to move the gene from pCU20 into pCU40 in BOTH directions in the same
reaction? How could you subsequently distinguish
the two variants?
Pst
I.
Can
use an enzyme such as Sma I that would produce different sized fragments in the
two orientations or Western blot
to look for protein, or PCR to determine orientation, or sequence the DNA, or
perform a Northern blot to look for mRNA.
8) 16 pts. Answer
this one or #9.
You are interested in two X-linked recessive lethal mutations in
Drosophila. One, lethal-1 (l-1), is known to map to 33.0 cM. The other l-2ts is a conditional (temperature-sensitive) lethal
that is inviable at high temperature. The marker genes cm–carmine eyes, and m--miniature wings map near 18 and 44 cM respectively.
To map 1-2ts you set up the following cross at high
temperature (30˚C):
cm l-2ts / l-1 m females X cm m /Y males.
The following progeny
phenotypes were observed:
Females: carmine 420
miniature 420
wild-type 80
carmine,
miniature 80
Males wild-type 5
Where does the l-2
gene map? (Show your work.) From one of your exams:
Assume 1000 males were produced, like the females, and
only 5 survived. Since all 5
lived, they must have arisen from a cross-over between l-2ts and l-1. When such a crossover occurs, half the
female gametes will be wild type, giving viable males.
This means 10/1000 males had this recombination or
1%. Therefore, 1 cM separates
1-2ts and l-1. l-2ts could be at
either 32.0 or 34.0 cM.
Considering the two possible orders:

It is obvious that l-2ts must be closer to cm and thus at 32.0 cM. If l-2ts were closer to m, the surviving males would
have been cm m.
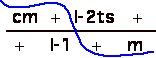
9) 16 pts. Answer this one or #8.
The
white gene is located near the
tip of the X-chromosome in Drosophila. In a large-scale
experiment (50,000 flies), wild-type males were gamma-irradiated and crossed to
white-eyed females. 15 red-eyed
males were isolated. 13 of these
males were sterile suggesting they lacked a Y chromosome. When the two fertile males were crossed
with white-eyed females one male produced 50% white and 50% red-eyed progeny
with no differences between the sexes, and the second produced all red-eyed
males and all white-eyed females.
Explain
how the 3 types of red-eyed males arose.
The 13 sterile males lacking their Y arose from
maternal non-disjunction. They
inherited their w+ X from their father and nothing from their
mother.
The first red-eyed male likely carried a translocation
of the w+ gene to an autosome. When crossed to females, half the progeny inherited the w+-bearing autosome and had red
eyes, while half inherited the wild-type autosome (non-translocation) and had
white eyes. The paternal X
chromosome was deleted for the white gene.
Figure
The second red-eyed male likely carried a
translocation of the w+ gene to the Y chromosome. When crossed to females, all the male progeny inherited the
Y-w+ chromosome and had red eyes. Their white-eyed sisters inherited the ∆w chromosome from their father
and a w- chromosome from their mothers.
Figure: