Abstract
Background Group 1 pulmonary arterial hypertension (PAH) is a lethal vasculopathy characterized by pathogenic remodeling of pulmonary arterioles leading to increased pulmonary pressures, right ventricular hypertrophy and heart failure. Recent high-throughput sequencing studies have identified additional PAH risk genes and suggested differences in genetic causes by age of onset. However, known risk genes explain only 15-20% of non-familial idiopathic PAH cases.
Methods To identify new risk genes, we utilized an international consortium of 4,241 PAH cases with 4,175 sequenced exomes (n=2,572 National Biological Sample and Data Repository for PAH; n=469 Columbia University Irving Medical Center, enriched for pediatric trios) and 1,134 sequenced genomes (UK NIHR Bioresource – Rare Diseases Study). Most of the cases were adult-onset disease (93%), and 55% idiopathic (IPAH) and 35% associated with other diseases (APAH). We identified protein-coding variants and performed rare variant association analyses in unrelated participants of European ancestry, including 2,789 cases and 18,819 controls (11,101 unaffected parents from the Simons Powering Autism Research for Knowledge study and 7,718 gnomAD individuals). We analyzed de novo variants in 124 pediatric trios.
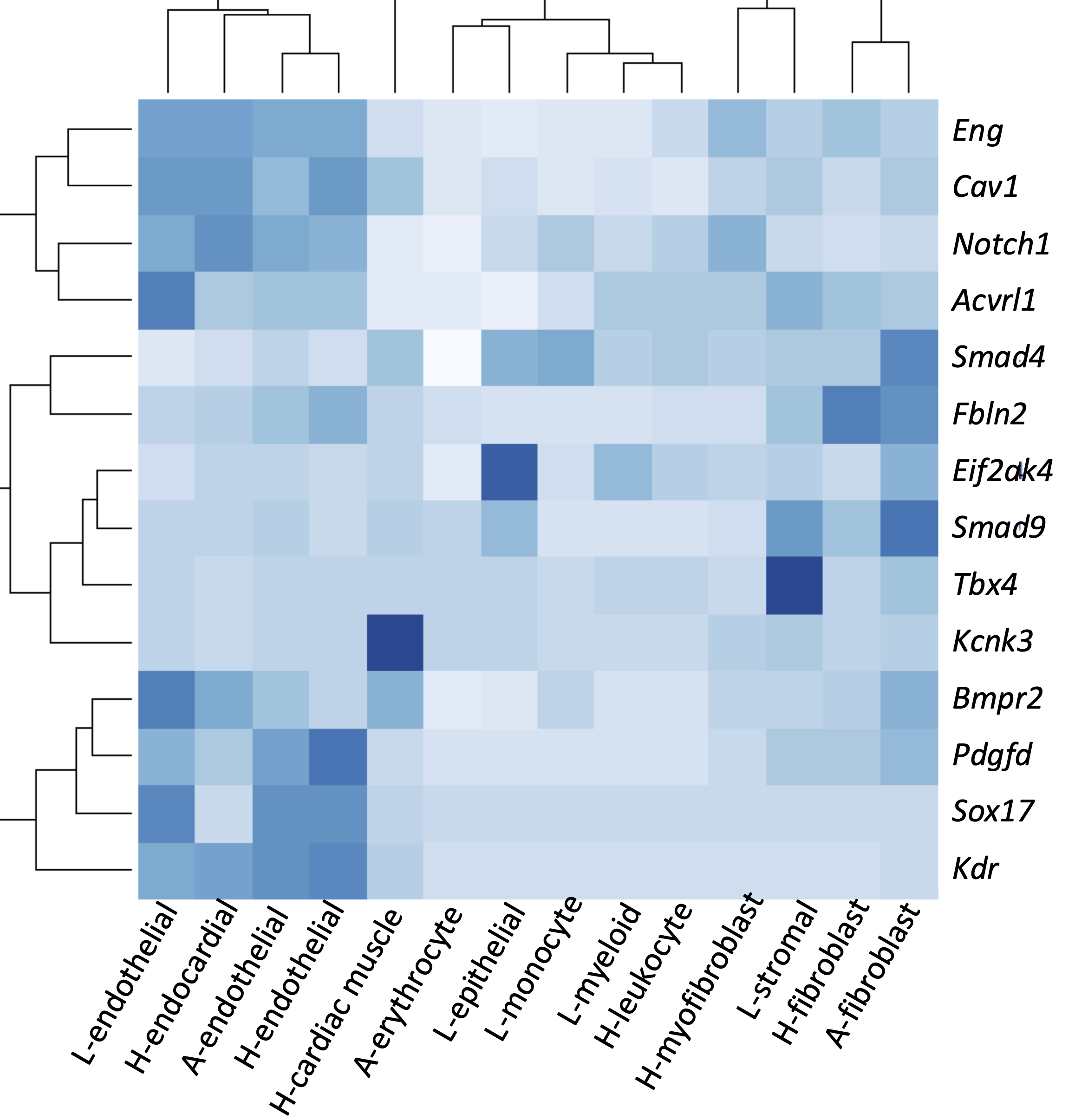
Results Seven genes with rare deleterious variants were significantly associated (false discovery rate <0.1) with IPAH, including three known genes (BMPR2, GDF2, and TBX4), two recently identified candidate genes (SOX17, KDR), and two new candidate genes (FBLN2, fibulin 2; PDGFD, platelet-derived growth factor D). The candidate genes exhibit expression patterns in lung and heart similar to that of known PAH risk genes, and most of the variants occur in conserved protein domains. Variants in known PAH gene, ACVRL1, showed association with APAH. Predicted deleterious de novo variants in pediatric cases exhibited a significant burden compared to the background mutation rate (2.5x, p=7.0E-6). At least eight novel candidate genes carrying de novo variants have plausible roles in lung/heart development.
Conclusions Rare variant analysis of a large international consortium identifies two new candidate genes - FBLN2 and PDGFD. The new genes have known functions in vasculogenesis and remodeling but have not been previously implicated in PAH. Trio analysis predicts that ~15% of pediatric IPAH may be explained by de novo variants.
A preprint of this paper is available on bioRxiv: https://doi.org/10.1101/2020.05.29.124255