JUSTIN SEONYONG LEE
JUSTIN
SEONYONG
LEE
Fine-Tuning SciBERT to Recognize Drug Names and Adverse Effects
November 2021
In this post, I go through a project I did for the Hugging Face (HF) Community Event
on November 15-19, 2021. I used the 🤗
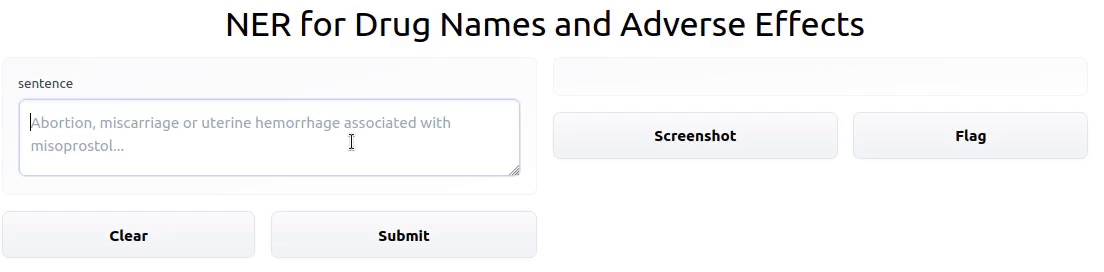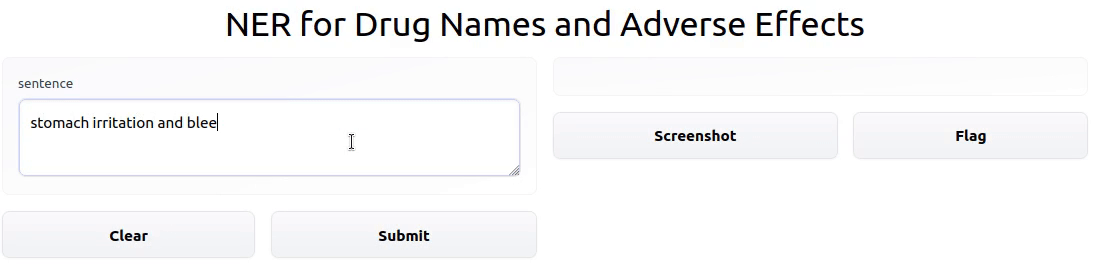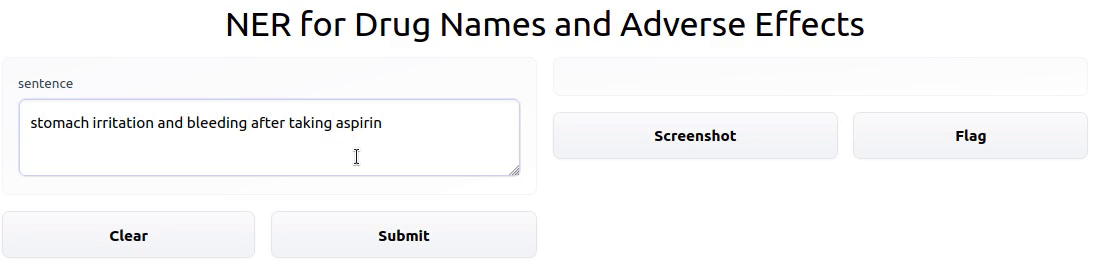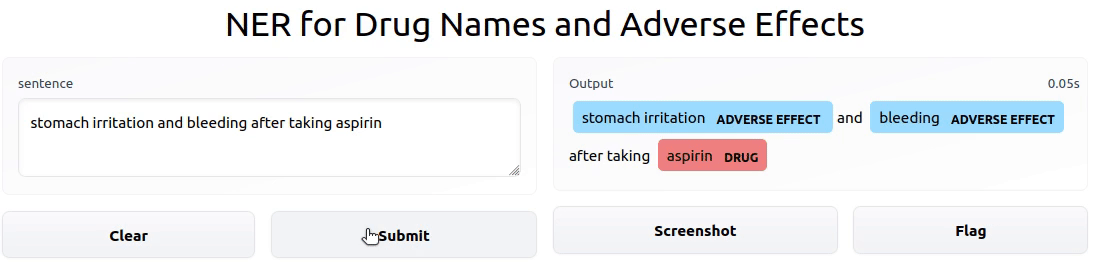
To showcase the final result, I deployed an interactive Gradio app powered by the model on HF Spaces.
 The HF ecosystem, particularly the
The HF ecosystem, particularly the
However, the data could not used as-is, because drugs and AEs are identified one at a time, as opposed to all at once for a given sentence. Therefore, if a unique sentence contains multiple drugs or AEs, that sentence would appear multiple times in the dataset. For example, below is a unique sentence appearing repeatedly in this manner:
In order to rectify this, I created a single set of labels for each sentence. I first grouped the rows of the dataset by sentence, then gathered all of the unique starting and ending indices for all drugs and AEs appearing in each sentence. Finally, I performed a single pass to tag all tokens in each sentence with the correct labeled entities. I followed the IOB tagging format to label each token. Since we have two entities ("drug" and "AE"), each token was assigned one of five possible labels:
•
•
•
•
•
This resulted in a new dataset of 4271 entries, with a total of 110,497 individual tokens after processing via the SciBERT tokenizer. The dataset was split 75-25 into training and test sets.
Once a model is pre-trained, developers and researchers are able to train it further on other tasks, also known as "fine-tuning." In this setting, I added a token classification layer to the pre-trained SciBERT model using the
After the model was trained, validation metrics were computed on the held-out test set:
The metrics were computed by the Trainer object using the
One interesting observation about the test set metrics is that the model performs better on
Once fine-tuning was complete, I uploaded the fine-tuned model to the HF model Hub. From there, anyone with the model name can import it into a
My final notebook from the event, which contains the data preprocessing pipeline outlined in this post, as well as the code to fine-tune the model, can be found on GitHub here.
Introduction to the CoNLL-2003 Shared Task: Language-Independent Named Entity Recognition, Eric F. Tjong Kim Sang & Fien De Meulder (2003)
transformers library to fine-tune the
allenai/scibert_scivocab_uncased
model
on the ade_corpus_v2
dataset.
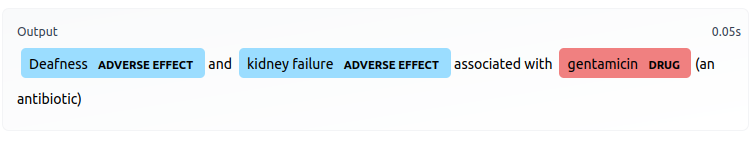
The fine-tuned model is able to perform Named Entity Recognition (NER) to label drug names and adverse drug
effects.
To showcase the final result, I deployed an interactive Gradio app powered by the model on HF Spaces.
Contents
Introduction [Top]
According to Wikipedia, an adverse effect (or "AE") is "an undesired harmful effect resulting from a medication or other intervention." The objective of this project was to train a machine learning model that tags adverse effects in an input text sample. The text could come from scientific publications, social media posts, drug labels, and other relevant sources. The HF ecosystem, particularly the
The HF ecosystem, particularly the transformers and datasets
libraries, made this task achievable in a matter of days and with low overhead. Model training
was done on a GPU-enabled
AWS p3.2xlarge instance, which was provided by AWS for the duration
of the Community Event.
Preparing the Dataset [Top]
The first task was to prepare a dataset that could be used to train an NER model. Theade_corpus_v2
dataset on the HF Hub was an excellent starting point, featuring (in the
Ade_corpus_v2_drug_ade_relation
subset) thousands of example texts with labeled spans for not only AEs, but also drug names.
Because of this, I was able to go beyond the original objective by training the model simultaneously to tag
drugs.
However, the data could not used as-is, because drugs and AEs are identified one at a time, as opposed to all at once for a given sentence. Therefore, if a unique sentence contains multiple drugs or AEs, that sentence would appear multiple times in the dataset. For example, below is a unique sentence appearing repeatedly in this manner:
{'text': 'After therapy for diabetic coma with insulin (containing the preservative cresol) and
electrolyte solutions was started, the patient complained of increasing myalgia, developed a high fever
and respiratory and metabolic acidosis and lost consciousness.', 'drug': 'insulin', 'effect':
'increasing myalgia', 'indexes': {'drug': {'start_char': [37], 'end_char': [44]}, 'effect':
{'start_char': [147], 'end_char': [165]}}}
{'text': 'After therapy for diabetic coma with insulin (containing the preservative cresol) and
electrolyte solutions was started, the patient complained of increasing myalgia, developed a high fever
and respiratory and metabolic acidosis and lost consciousness.', 'drug': 'cresol', 'effect': 'lost
consciousness', 'indexes': {'drug': {'start_char': [74], 'end_char': [80]}, 'effect': {'start_char':
[233], 'end_char': [251]}}}
{'text': 'After therapy for diabetic coma with insulin (containing the preservative cresol) and
electrolyte solutions was started, the patient complained of increasing myalgia, developed a high fever
and respiratory and metabolic acidosis and lost consciousness.', 'drug': 'cresol', 'effect': 'high
fever', 'indexes': {'drug': {'start_char': [74], 'end_char': [80]}, 'effect': {'start_char': [179],
'end_char': [189]}}}
>
Therefore, despite containing 6821 rows, the dataset only contains around 4200 unique text samples.
Having text samples repeat in this manner is problematic in an NER setting -
if we treat each row as a unique datapoint, then we would confuse the model with
contradictory labels for the same entities. For instance, the above example would be annotated like below :
After therapy for diabetic coma with <DRUG>insulin</DRUG> (containing the preservative cresol) and electrolyte
solutions was started, the patient complained of <EFFECT>increasing
myalgia</EFFECT>, developed a high fever and respiratory and
metabolic acidosis and lost consciousness.
After therapy for diabetic coma with insulin (containing the preservative <DRUG>cresol</DRUG>) and
electrolyte solutions was started, the patient complained of increasing myalgia, developed a high fever
and respiratory and metabolic acidosis and <EFFECT>lost
consciousness</EFFECT>.
In the first instance of the sentence, "insulin" is labeled as a drug, and "increasing myalgia" as an AE. In
the second
instance, neither of the entities from the first instance are labeled, and instead, "cresol" is labeled as a
drug and "lost consciousness" as an entity.
Therefore, if the model learns to label "insulin" as a drug after seeing the first instance, it would
actually be penalized for doing so
in the second instance, where "insulin" is not assigned a label at all.
In order to rectify this, I created a single set of labels for each sentence. I first grouped the rows of the dataset by sentence, then gathered all of the unique starting and ending indices for all drugs and AEs appearing in each sentence. Finally, I performed a single pass to tag all tokens in each sentence with the correct labeled entities. I followed the IOB tagging format to label each token. Since we have two entities ("drug" and "AE"), each token was assigned one of five possible labels:
•
B-DRUG - the beginning of a drug entity
•
I-DRUG - inside a drug entity
•
B-EFFECT - the beginning of an AE entity
•
I-EFFECT - inside an AE entity
•
O - outside any entity being tagged
This resulted in a new dataset of 4271 entries, with a total of 110,497 individual tokens after processing via the SciBERT tokenizer. The dataset was split 75-25 into training and test sets.
Fine-Tuning SciBERT [Top]
SciBERT is a pre-trained BERT model released by the Allen Institute for AI. It was specifically pre-trained on a large corpus of scientific publications. Pre-training a model entails training it on an objective designed to make the model learn the relationships between tokens in the training data. If the pre-training corpus comes from a specific genre (e.g. scientific publications, social media posts, source code, specific languages), then the model would learn the particulars of that genre.Once a model is pre-trained, developers and researchers are able to train it further on other tasks, also known as "fine-tuning." In this setting, I added a token classification layer to the pre-trained SciBERT model using the
AutoModelForTokenClassification class.
Unlike pre-training, which is expensive, time-consuming, and requires a lot of data,
fine-tuning on a well-aligned task is often a quick process.
Here, the model only needed 3 epochs to reach a reasonable
test set performance.
After the model was trained, validation metrics were computed on the held-out test set:
| Category | Precision | Recall | F1 | Count |
DRUG |
0.923 | 0.966 | 0.944 | 1299* |
EFFECT |
0.805 | 0.873 | 0.838 | 1412* |
Overall Tokens |
0.861 | 0.918 | 0.888 | 27759 |
* These counts and their corresponding metrics are over entities, as opposed to tokens. The counts
exactly correspond to the
number of tokens of the given type with the prefix B-. By contrast,
the third row is describing performance over tokens, including the non-entity class O.
|
||||
The metrics were computed by the Trainer object using the
seqeval Python package. seqeval
is a re-implementation of the popular conlleval Perl script, which evaluates NER
performance
according to the specifications in Tjong Kim Sang & Buchholz (2000).
It is important to note that the metrics are computed on an entity-level and not on a token-level,
meaning (emphasis mine):
[p]recision is the percentage of named entities found by the learning system that are correct. Recall is the percentage of named entities present in the corpus that are found by the system. A named entity is correct only if it is an exact match of the corresponding entity in the data file (Tjong Kim Sang & De Meulder 2003).Thus, it does not suffice for the model to correctly label part of an entity - it must correctly label all and only those tokens that comprise an entity in order to receive credit.
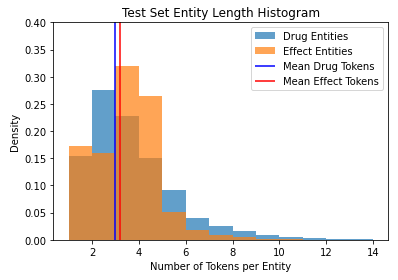
One interesting observation about the test set metrics is that the model performs better on
DRUG entities
than on
EFFECT entities, despite there being more examples of effects than drugs in
our training set. My guess is this is because commercial drug names tend to follow
a formula and
are thus
synthetically
generated, whereas adverse effects rely on words that only indicate an adverse effect in particular
contexts. As we can
see below, the length of the average DRUG entity in our test set is slightly
shorter than that of the average
EFFECT entity, although the latter skews right a bit more:

Once fine-tuning was complete, I uploaded the fine-tuned model to the HF model Hub. From there, anyone with the model name can import it into a
pipeline object to
perform
inference on
custom input:
from transformers import (AutoModelForTokenClassification,
AutoTokenizer,
pipeline,
)
model_checkpoint = "jsylee/scibert_scivocab_uncased-finetuned-ner"
model = AutoModelForTokenClassification.from_pretrained(model_checkpoint, num_labels=5,
id2label={0: 'O', 1: 'B-DRUG', 2: 'I-DRUG', 3: 'B-EFFECT', 4: 'I-EFFECT'}
)
tokenizer = AutoTokenizer.from_pretrained(model_checkpoint)
model_pipeline = pipeline(task="ner", model=model, tokenizer=tokenizer)
print( model_pipeline ("Abortion, miscarriage or uterine hemorrhage associated with misoprostol (Cytotec), a labor-inducing drug."))
Conclusion [Top]
As a participant in the recent HF Community Event for the second part of their Transformer course, I modified a dataset of medical text and used it to fine-tune SciBERT on an NER task. To me, this experience demonstrated how easy HF has made it to train and share custom Transformer-based models for natural language processing. It also showed how pre-training vastly scales up the application of Transformer-based models in practice, by making it feasible to train high-performing models across a variety of NLP objectives at relatively low cost.My final notebook from the event, which contains the data preprocessing pipeline outlined in this post, as well as the code to fine-tune the model, can be found on GitHub here.

References [Top]
Introduction to the CoNLL-2000 Shared Task: Chunking, Eric F. Tjong Kim Sang & Sabine Buchholz (2000)Introduction to the CoNLL-2003 Shared Task: Language-Independent Named Entity Recognition, Eric F. Tjong Kim Sang & Fien De Meulder (2003)