Abstract
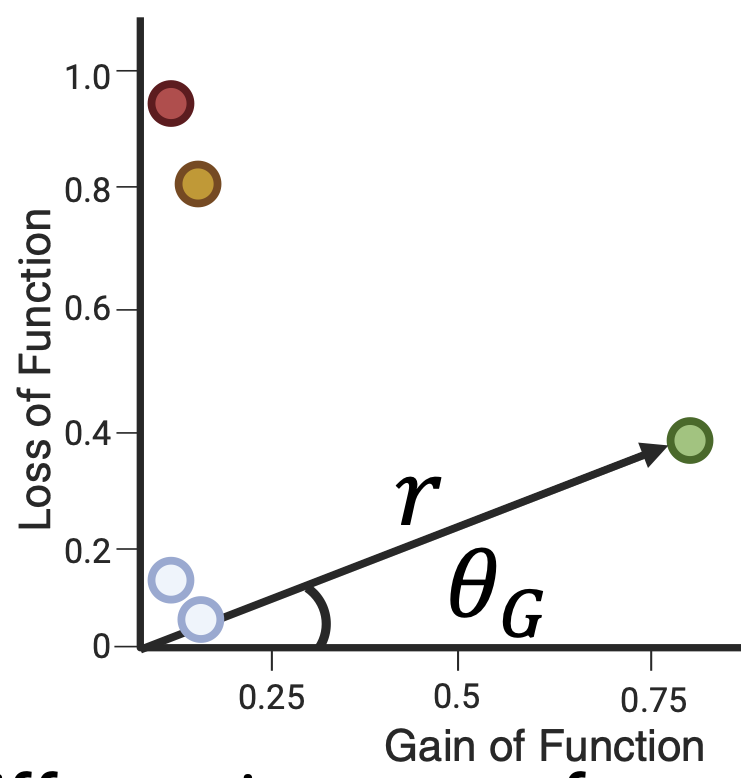
Accurate prediction of the functional impact of missense variants is fundamentally important for disease gene discovery, clinical genetic diagnostics, therapeutic strategies, and protein engineering. Previous efforts have focused on predicting a binary pathogenicity classification, but the functional impact of missense variants is multi-dimensional. Pathogenic missense variants in the same gene may act through different modes of action (i.e., gain/loss-of-function) by affecting multiple protein biochemical properties. They may result in distinct clinical conditions that require different treatments. We developed a new method, PreMode, to perform gene-specific mode-of-action predictions. PreMode models effects of coding sequence variants using SE(3)-equivariant graph neural networks on protein sequences and structures. Using the largest-to-date set of mode-of-action-labeled missense variants, we show that PreMode reaches state-of-the-art performance in multiple types of mode-of-action predictions by efficient transfer-learning. Additionally, PreMode prediction of G/LoF variants in a kinase is consistent with inactive-active conformation transition. Finally, we show that PreMode enables improved mutagenesis analysis, clinical diagnosis and more broadly, artificial GoF engineering of proteins.